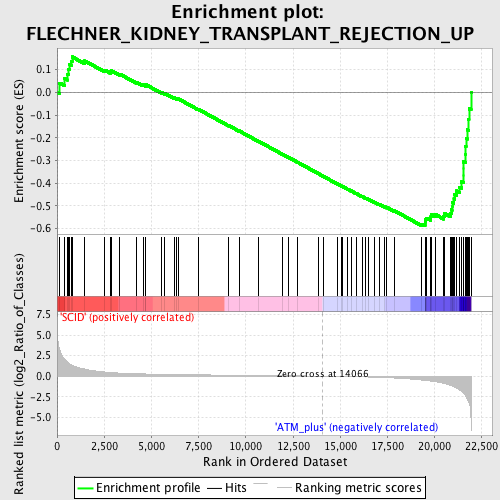
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
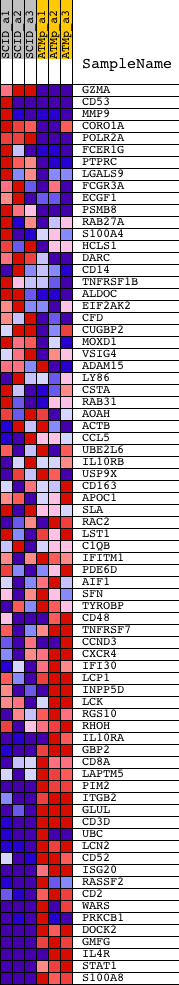
| GeneSet | FLECHNER_KIDNEY_TRANSPLANT_REJECTION_UP |
| Enrichment Score (ES) | -0.58871233 |
| Normalized Enrichment Score (NES) | -1.7126247 |
| Nominal p-value | 0.003960396 |
| FDR q-value | 0.16237326 |
| FWER p-Value | 0.863 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | GZMA | 1417898_a_at | 140 | 3.145 | 0.0408 | No | ||
| 2 | CD53 | 1439589_at 1448617_at 1459744_at | 381 | 2.091 | 0.0612 | No | ||
| 3 | MMP9 | 1416298_at 1448291_at | 529 | 1.771 | 0.0810 | No | ||
| 4 | CORO1A | 1416246_a_at 1435288_at 1455269_a_at | 597 | 1.629 | 0.1024 | No | ||
| 5 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 664 | 1.498 | 0.1219 | No | ||
| 6 | FCER1G | 1418340_at | 768 | 1.376 | 0.1378 | No | ||
| 7 | PTPRC | 1422124_a_at 1440165_at | 795 | 1.337 | 0.1567 | No | ||
| 8 | LGALS9 | 1421217_a_at | 1469 | 0.853 | 0.1387 | No | ||
| 9 | FCGR3A | 1425225_at 1448620_at | 2523 | 0.504 | 0.0981 | No | ||
| 10 | ECGF1 | 1432181_s_at | 2834 | 0.446 | 0.0906 | No | ||
| 11 | PSMB8 | 1422962_a_at 1444619_x_at | 2873 | 0.439 | 0.0954 | No | ||
| 12 | RAB27A | 1425284_a_at 1425285_a_at 1429123_at 1454438_at | 3322 | 0.372 | 0.0805 | No | ||
| 13 | S100A4 | 1424542_at | 4206 | 0.295 | 0.0446 | No | ||
| 14 | HCLS1 | 1418842_at | 4553 | 0.273 | 0.0328 | No | ||
| 15 | DARC | 1432273_a_at | 4676 | 0.267 | 0.0313 | No | ||
| 16 | CD14 | 1417268_at | 4702 | 0.266 | 0.0341 | No | ||
| 17 | TNFRSF1B | 1418099_at 1448951_at | 5527 | 0.230 | -0.0001 | No | ||
| 18 | ALDOC | 1424714_at 1451461_a_at | 5701 | 0.223 | -0.0047 | No | ||
| 19 | EIF2AK2 | 1422005_at 1422006_at 1440866_at | 6192 | 0.205 | -0.0240 | No | ||
| 20 | CFD | 1417867_at | 6323 | 0.201 | -0.0270 | No | ||
| 21 | CUGBP2 | 1423895_a_at 1440322_at 1441072_at 1443545_at 1444152_at 1444533_at 1445306_at 1446670_at 1450069_a_at 1451154_a_at 1451155_at 1457553_at 1458263_at | 6427 | 0.197 | -0.0287 | No | ||
| 22 | MOXD1 | 1422643_at | 7492 | 0.164 | -0.0749 | No | ||
| 23 | VSIG4 | 1451651_at | 9095 | 0.122 | -0.1464 | No | ||
| 24 | ADAM15 | 1416080_at 1425170_a_at 1438760_x_at 1454206_a_at | 9641 | 0.109 | -0.1697 | No | ||
| 25 | LY86 | 1422903_at | 10655 | 0.085 | -0.2147 | No | ||
| 26 | CSTA | 1435760_at | 11937 | 0.054 | -0.2725 | No | ||
| 27 | RAB31 | 1416165_at 1431691_a_at | 12278 | 0.046 | -0.2874 | No | ||
| 28 | AOAH | 1450764_at 1460156_at | 12738 | 0.035 | -0.3078 | No | ||
| 29 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 13833 | 0.006 | -0.3578 | No | ||
| 30 | CCL5 | 1418126_at | 14085 | -0.001 | -0.3693 | No | ||
| 31 | UBE2L6 | 1417172_at 1447633_x_at | 14863 | -0.025 | -0.4044 | No | ||
| 32 | IL10RB | 1419455_at | 15061 | -0.032 | -0.4130 | No | ||
| 33 | USP9X | 1420922_at 1420923_at 1428193_at 1428194_at 1450037_at 1450038_s_at 1450039_at | 15138 | -0.034 | -0.4159 | No | ||
| 34 | CD163 | 1419144_at | 15369 | -0.044 | -0.4258 | No | ||
| 35 | APOC1 | 1417561_at | 15565 | -0.051 | -0.4340 | No | ||
| 36 | SLA | 1420818_at 1420819_at 1441761_at 1447813_x_at | 15852 | -0.064 | -0.4461 | No | ||
| 37 | RAC2 | 1417620_at 1440208_at | 16178 | -0.079 | -0.4598 | No | ||
| 38 | LST1 | 1425548_a_at | 16332 | -0.087 | -0.4655 | No | ||
| 39 | C1QB | 1417063_at 1434366_x_at 1437726_x_at 1457322_at | 16476 | -0.097 | -0.4705 | No | ||
| 40 | IFITM1 | 1424254_at | 16803 | -0.119 | -0.4837 | No | ||
| 41 | PDE6D | 1416843_at | 17051 | -0.138 | -0.4929 | No | ||
| 42 | AIF1 | 1418204_s_at | 17325 | -0.164 | -0.5029 | No | ||
| 43 | SFN | 1448612_at | 17471 | -0.178 | -0.5069 | No | ||
| 44 | TYROBP | 1450792_at | 17866 | -0.220 | -0.5216 | No | ||
| 45 | CD48 | 1427301_at | 19317 | -0.467 | -0.5809 | No | ||
| 46 | TNFRSF7 | 1452389_at | 19488 | -0.507 | -0.5811 | Yes | ||
| 47 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 19489 | -0.507 | -0.5735 | Yes | ||
| 48 | CXCR4 | 1448710_at | 19495 | -0.509 | -0.5661 | Yes | ||
| 49 | IFI30 | 1422476_at | 19514 | -0.517 | -0.5592 | Yes | ||
| 50 | LCP1 | 1415983_at 1448160_at | 19572 | -0.530 | -0.5538 | Yes | ||
| 51 | INPP5D | 1418110_a_at 1424195_a_at | 19788 | -0.590 | -0.5548 | Yes | ||
| 52 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 19798 | -0.593 | -0.5463 | Yes | ||
| 53 | RGS10 | 1416882_at | 19812 | -0.598 | -0.5379 | Yes | ||
| 54 | RHOH | 1429319_at 1441385_at 1443015_at | 20029 | -0.669 | -0.5378 | Yes | ||
| 55 | IL10RA | 1437808_x_at 1448731_at 1456173_at | 20476 | -0.860 | -0.5453 | Yes | ||
| 56 | GBP2 | 1418240_at 1435906_x_at | 20519 | -0.880 | -0.5340 | Yes | ||
| 57 | CD8A | 1440164_x_at 1440811_x_at | 20843 | -1.091 | -0.5324 | Yes | ||
| 58 | LAPTM5 | 1426025_s_at 1436905_x_at 1447742_at 1459841_x_at | 20894 | -1.142 | -0.5175 | Yes | ||
| 59 | PIM2 | 1417216_at | 20918 | -1.164 | -0.5011 | Yes | ||
| 60 | ITGB2 | 1450678_at | 20924 | -1.167 | -0.4838 | Yes | ||
| 61 | GLUL | 1426235_a_at 1426236_a_at | 21020 | -1.262 | -0.4692 | Yes | ||
| 62 | CD3D | 1422828_at | 21050 | -1.299 | -0.4511 | Yes | ||
| 63 | UBC | 1420494_x_at 1425965_at 1425966_x_at 1432827_x_at 1437666_x_at 1454373_x_at | 21150 | -1.420 | -0.4343 | Yes | ||
| 64 | LCN2 | 1427747_a_at | 21308 | -1.632 | -0.4170 | Yes | ||
| 65 | CD52 | 1460218_at | 21414 | -1.824 | -0.3944 | Yes | ||
| 66 | ISG20 | 1419569_a_at | 21521 | -2.074 | -0.3681 | Yes | ||
| 67 | RASSF2 | 1428392_at 1444889_at | 21522 | -2.075 | -0.3370 | Yes | ||
| 68 | CD2 | 1418770_at | 21523 | -2.075 | -0.3059 | Yes | ||
| 69 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 21628 | -2.373 | -0.2750 | Yes | ||
| 70 | PRKCB1 | 1423478_at 1438981_at 1443144_at 1459674_at 1460419_a_at | 21636 | -2.405 | -0.2393 | Yes | ||
| 71 | DOCK2 | 1422808_s_at 1437282_at 1438334_at 1459382_at | 21660 | -2.487 | -0.2030 | Yes | ||
| 72 | GMFG | 1419193_a_at 1419194_s_at | 21717 | -2.780 | -0.1639 | Yes | ||
| 73 | IL4R | 1421034_a_at 1423996_a_at 1447858_x_at | 21800 | -3.148 | -0.1204 | Yes | ||
| 74 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 21816 | -3.247 | -0.0723 | Yes | ||
| 75 | S100A8 | 1419394_s_at | 21925 | -5.183 | 0.0005 | Yes |