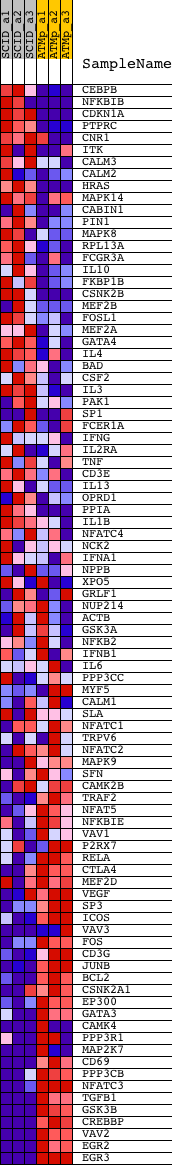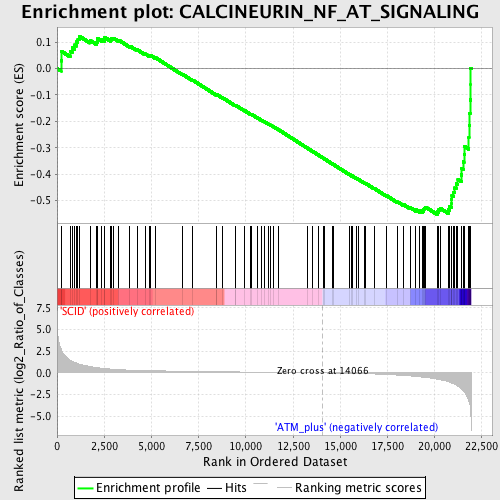
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CALCINEURIN_NF_AT_SIGNALING |
| Enrichment Score (ES) | -0.5522046 |
| Normalized Enrichment Score (NES) | -1.662017 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.20067726 |
| FWER p-Value | 0.984 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 1418901_at 1427844_a_at | 208 | 2.772 | 0.0290 | No | ||
| 2 | NFKBIB | 1421266_s_at 1446718_at | 224 | 2.680 | 0.0655 | No | ||
| 3 | CDKN1A | 1421679_a_at 1424638_at | 689 | 1.463 | 0.0646 | No | ||
| 4 | PTPRC | 1422124_a_at 1440165_at | 795 | 1.337 | 0.0784 | No | ||
| 5 | CNR1 | 1419425_at 1434172_at | 930 | 1.208 | 0.0890 | No | ||
| 6 | ITK | 1417171_at 1430833_at 1452518_a_at 1456836_at 1457120_at | 1018 | 1.129 | 0.1007 | No | ||
| 7 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 1103 | 1.061 | 0.1116 | No | ||
| 8 | CALM2 | 1422414_a_at 1423807_a_at | 1171 | 1.010 | 0.1225 | No | ||
| 9 | HRAS | 1422407_s_at 1424132_at | 1741 | 0.725 | 0.1066 | No | ||
| 10 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 2079 | 0.612 | 0.0996 | No | ||
| 11 | CABIN1 | 1433686_at 1437794_at 1440596_at | 2121 | 0.599 | 0.1061 | No | ||
| 12 | PIN1 | 1416228_at | 2127 | 0.598 | 0.1142 | No | ||
| 13 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 2363 | 0.538 | 0.1109 | No | ||
| 14 | RPL13A | 1417608_a_at 1433928_a_at 1455001_x_at 1455485_x_at | 2521 | 0.504 | 0.1107 | No | ||
| 15 | FCGR3A | 1425225_at 1448620_at | 2523 | 0.504 | 0.1177 | No | ||
| 16 | IL10 | 1450330_at | 2846 | 0.444 | 0.1091 | No | ||
| 17 | FKBP1B | 1449429_at | 2878 | 0.438 | 0.1137 | No | ||
| 18 | CSNK2B | 1416728_at | 3005 | 0.419 | 0.1138 | No | ||
| 19 | MEF2B | 1421541_a_at | 3255 | 0.381 | 0.1077 | No | ||
| 20 | FOSL1 | 1417487_at 1417488_at | 3849 | 0.321 | 0.0850 | No | ||
| 21 | MEF2A | 1421252_a_at 1425426_a_at 1427185_at 1427186_a_at 1452347_at | 4244 | 0.292 | 0.0710 | No | ||
| 22 | GATA4 | 1418863_at 1418864_at 1441364_at | 4673 | 0.268 | 0.0552 | No | ||
| 23 | IL4 | 1449864_at | 4912 | 0.257 | 0.0478 | No | ||
| 24 | BAD | 1416582_a_at 1416583_at | 4939 | 0.255 | 0.0502 | No | ||
| 25 | CSF2 | 1427429_at | 5226 | 0.242 | 0.0405 | No | ||
| 26 | IL3 | 1450566_at | 6633 | 0.190 | -0.0213 | No | ||
| 27 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 7172 | 0.174 | -0.0435 | No | ||
| 28 | SP1 | 1418180_at 1448994_at 1454852_at | 8439 | 0.138 | -0.0995 | No | ||
| 29 | FCER1A | 1421775_at | 8449 | 0.138 | -0.0980 | No | ||
| 30 | IFNG | 1425947_at | 8785 | 0.130 | -0.1115 | No | ||
| 31 | IL2RA | 1420691_at 1420692_at | 9422 | 0.113 | -0.1391 | No | ||
| 32 | TNF | 1419607_at | 9455 | 0.113 | -0.1390 | No | ||
| 33 | CD3E | 1422105_at 1445748_at | 9934 | 0.101 | -0.1594 | No | ||
| 34 | IL13 | 1420802_at | 10244 | 0.094 | -0.1723 | No | ||
| 35 | OPRD1 | 1422121_at | 10321 | 0.092 | -0.1745 | No | ||
| 36 | PPIA | 1417451_a_at | 10595 | 0.086 | -0.1858 | No | ||
| 37 | IL1B | 1449399_a_at | 10824 | 0.081 | -0.1951 | No | ||
| 38 | NFATC4 | 1423380_s_at 1432821_at 1454369_a_at | 10981 | 0.077 | -0.2012 | No | ||
| 39 | NCK2 | 1416796_at 1416797_at 1458432_at | 11200 | 0.072 | -0.2101 | No | ||
| 40 | IFNA1 | 1450564_x_at | 11304 | 0.070 | -0.2139 | No | ||
| 41 | NPPB | 1450791_at | 11469 | 0.066 | -0.2205 | No | ||
| 42 | XPO5 | 1428560_at 1451468_s_at | 11737 | 0.059 | -0.2319 | No | ||
| 43 | GRLF1 | 1434009_at 1440675_at 1442181_at | 13278 | 0.021 | -0.3021 | No | ||
| 44 | NUP214 | 1434351_at 1456503_at 1457815_at | 13511 | 0.015 | -0.3125 | No | ||
| 45 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 13833 | 0.006 | -0.3271 | No | ||
| 46 | GSK3A | 1435638_at | 14111 | -0.002 | -0.3397 | No | ||
| 47 | NFKB2 | 1425902_a_at 1429128_x_at | 14187 | -0.004 | -0.3431 | No | ||
| 48 | IFNB1 | 1422305_at | 14608 | -0.017 | -0.3621 | No | ||
| 49 | IL6 | 1450297_at | 14615 | -0.017 | -0.3622 | No | ||
| 50 | PPP3CC | 1420743_a_at 1430025_at | 15477 | -0.048 | -0.4009 | No | ||
| 51 | MYF5 | 1420757_at | 15613 | -0.052 | -0.4064 | No | ||
| 52 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 15628 | -0.053 | -0.4063 | No | ||
| 53 | SLA | 1420818_at 1420819_at 1441761_at 1447813_x_at | 15852 | -0.064 | -0.4156 | No | ||
| 54 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 15972 | -0.069 | -0.4201 | No | ||
| 55 | TRPV6 | 1419615_at | 16299 | -0.085 | -0.4338 | No | ||
| 56 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 16317 | -0.086 | -0.4334 | No | ||
| 57 | MAPK9 | 1421876_at 1421877_at 1421878_at | 16802 | -0.119 | -0.4539 | No | ||
| 58 | SFN | 1448612_at | 17471 | -0.178 | -0.4820 | No | ||
| 59 | CAMK2B | 1448676_at 1455869_at | 18040 | -0.244 | -0.5046 | No | ||
| 60 | TRAF2 | 1451233_at | 18355 | -0.288 | -0.5150 | No | ||
| 61 | NFAT5 | 1426029_a_at 1438999_a_at 1439328_at 1439805_at 1448966_a_at 1451921_a_at 1458213_at | 18708 | -0.338 | -0.5264 | No | ||
| 62 | NFKBIE | 1431843_a_at 1458299_s_at | 18998 | -0.394 | -0.5342 | No | ||
| 63 | VAV1 | 1422932_a_at | 19214 | -0.442 | -0.5379 | No | ||
| 64 | P2RX7 | 1419853_a_at 1422218_at 1439787_at | 19351 | -0.474 | -0.5375 | No | ||
| 65 | RELA | 1419536_a_at | 19403 | -0.485 | -0.5331 | No | ||
| 66 | CTLA4 | 1419334_at | 19438 | -0.493 | -0.5278 | No | ||
| 67 | MEF2D | 1421388_at 1434487_at 1436642_x_at 1437300_at 1458901_at | 19516 | -0.517 | -0.5241 | No | ||
| 68 | VEGF | 1420909_at 1451959_a_at | 20130 | -0.707 | -0.5424 | Yes | ||
| 69 | SP3 | 1431804_a_at 1459360_at | 20191 | -0.732 | -0.5350 | Yes | ||
| 70 | ICOS | 1421930_at 1421931_at 1436598_at | 20279 | -0.774 | -0.5282 | Yes | ||
| 71 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20714 | -1.006 | -0.5341 | Yes | ||
| 72 | FOS | 1423100_at | 20795 | -1.059 | -0.5231 | Yes | ||
| 73 | CD3G | 1419178_at | 20877 | -1.120 | -0.5112 | Yes | ||
| 74 | JUNB | 1415899_at | 20884 | -1.130 | -0.4958 | Yes | ||
| 75 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 20911 | -1.157 | -0.4809 | Yes | ||
| 76 | CSNK2A1 | 1419034_at 1419035_s_at 1419036_at 1419037_at 1419038_a_at 1453427_at | 21003 | -1.246 | -0.4678 | Yes | ||
| 77 | EP300 | 1434765_at | 21052 | -1.301 | -0.4519 | Yes | ||
| 78 | GATA3 | 1448886_at | 21135 | -1.401 | -0.4362 | Yes | ||
| 79 | CAMK4 | 1421941_at 1426167_a_at 1439843_at 1442822_at 1452572_at | 21229 | -1.528 | -0.4192 | Yes | ||
| 80 | PPP3R1 | 1421786_at 1433591_at 1450368_a_at | 21405 | -1.812 | -0.4021 | Yes | ||
| 81 | MAP2K7 | 1421416_at 1425393_a_at 1425512_at 1425513_at 1440442_at 1451736_a_at 1457182_at | 21434 | -1.870 | -0.3774 | Yes | ||
| 82 | CD69 | 1428735_at | 21507 | -2.047 | -0.3523 | Yes | ||
| 83 | PPP3CB | 1427468_at 1428473_at 1428474_at 1433835_at 1446149_at 1459378_at | 21553 | -2.158 | -0.3244 | Yes | ||
| 84 | NFATC3 | 1419976_s_at 1452497_a_at | 21574 | -2.204 | -0.2947 | Yes | ||
| 85 | TGFB1 | 1420653_at 1445360_at | 21809 | -3.227 | -0.2606 | Yes | ||
| 86 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21817 | -3.254 | -0.2157 | Yes | ||
| 87 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21856 | -3.508 | -0.1687 | Yes | ||
| 88 | VAV2 | 1421272_at 1435065_x_at 1435244_at 1458758_at | 21875 | -3.671 | -0.1186 | Yes | ||
| 89 | EGR2 | 1427682_a_at 1427683_at | 21904 | -4.293 | -0.0602 | Yes | ||
| 90 | EGR3 | 1421486_at 1436329_at | 21908 | -4.438 | 0.0013 | Yes |