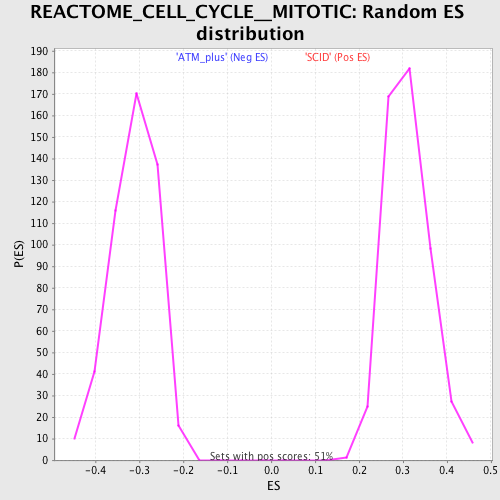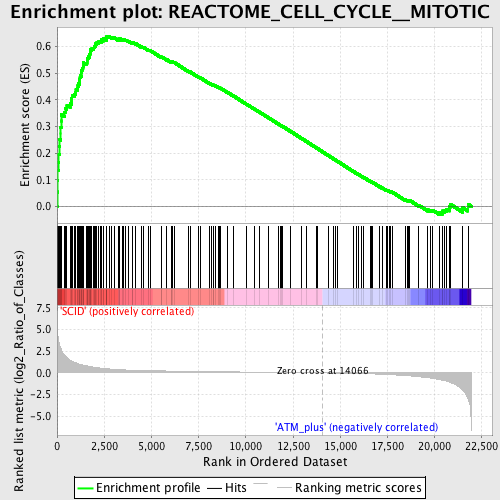
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.63936114 |
| Normalized Enrichment Score (NES) | 2.0564177 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 8 | 6.245 | 0.0542 | Yes | ||
| 2 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 24 | 4.996 | 0.0971 | Yes | ||
| 3 | POLE | 1441854_at 1448650_a_at | 38 | 4.383 | 0.1348 | Yes | ||
| 4 | CENPA | 1441864_x_at 1444416_at 1450842_a_at | 70 | 3.737 | 0.1660 | Yes | ||
| 5 | CCNB1 | 1419943_s_at 1419944_at | 88 | 3.586 | 0.1965 | Yes | ||
| 6 | SDCCAG8 | 1431203_at 1444831_at 1453946_a_at 1456538_at | 118 | 3.318 | 0.2242 | Yes | ||
| 7 | POLA2 | 1425794_at 1448369_at 1456713_at | 144 | 3.131 | 0.2504 | Yes | ||
| 8 | ZWINT | 1423724_at 1427539_a_at 1427540_at 1429786_a_at 1429787_x_at 1444717_at 1455382_at | 195 | 2.844 | 0.2729 | Yes | ||
| 9 | FEN1 | 1421731_a_at 1436454_x_at | 196 | 2.841 | 0.2977 | Yes | ||
| 10 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 214 | 2.722 | 0.3207 | Yes | ||
| 11 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 220 | 2.689 | 0.3439 | Yes | ||
| 12 | NEK2 | 1417299_at 1437579_at 1437580_s_at 1443999_at | 399 | 2.043 | 0.3536 | Yes | ||
| 13 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 453 | 1.925 | 0.3680 | Yes | ||
| 14 | MCM5 | 1415945_at 1436808_x_at | 521 | 1.778 | 0.3805 | Yes | ||
| 15 | CDKN1A | 1421679_a_at 1424638_at | 689 | 1.463 | 0.3856 | Yes | ||
| 16 | WEE1 | 1416773_at 1416774_at | 753 | 1.388 | 0.3948 | Yes | ||
| 17 | GMNN | 1417506_at | 766 | 1.376 | 0.4063 | Yes | ||
| 18 | RFC4 | 1424321_at 1438161_s_at | 788 | 1.350 | 0.4171 | Yes | ||
| 19 | LIG1 | 1416641_at | 921 | 1.215 | 0.4216 | Yes | ||
| 20 | PSMB9 | 1450696_at | 978 | 1.164 | 0.4292 | Yes | ||
| 21 | PSMD7 | 1432820_at 1451056_at 1454368_at | 999 | 1.147 | 0.4383 | Yes | ||
| 22 | RFC3 | 1423700_at 1432538_a_at | 1086 | 1.072 | 0.4437 | Yes | ||
| 23 | RFC5 | 1452917_at | 1102 | 1.062 | 0.4523 | Yes | ||
| 24 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 1125 | 1.041 | 0.4604 | Yes | ||
| 25 | KIF20A | 1449207_a_at | 1178 | 1.006 | 0.4668 | Yes | ||
| 26 | POLE2 | 1427094_at 1431440_at | 1190 | 1.000 | 0.4750 | Yes | ||
| 27 | NUP43 | 1432187_at 1432188_s_at | 1211 | 0.986 | 0.4827 | Yes | ||
| 28 | POLD1 | 1448187_at 1456055_x_at | 1223 | 0.980 | 0.4908 | Yes | ||
| 29 | BIRC5 | 1424278_a_at | 1283 | 0.944 | 0.4963 | Yes | ||
| 30 | PMF1 | 1416707_a_at 1438173_x_at | 1284 | 0.944 | 0.5046 | Yes | ||
| 31 | RPA2 | 1416433_at 1454011_a_at | 1307 | 0.930 | 0.5117 | Yes | ||
| 32 | CDC25B | 1421963_a_at | 1325 | 0.924 | 0.5190 | Yes | ||
| 33 | CCNB2 | 1450920_at | 1372 | 0.898 | 0.5247 | Yes | ||
| 34 | DHFR | 1419172_at 1430750_at | 1396 | 0.890 | 0.5314 | Yes | ||
| 35 | CDC45L | 1416575_at 1457838_at | 1397 | 0.889 | 0.5392 | Yes | ||
| 36 | PSMA5 | 1424681_a_at 1434356_a_at | 1570 | 0.805 | 0.5383 | Yes | ||
| 37 | CLASP2 | 1424836_a_at 1427328_a_at 1429976_at 1440802_at 1441726_at 1443320_at 1456911_at | 1584 | 0.799 | 0.5447 | Yes | ||
| 38 | KIF23 | 1450827_at 1453748_a_at 1455990_at | 1587 | 0.798 | 0.5515 | Yes | ||
| 39 | POLD2 | 1448277_at | 1620 | 0.785 | 0.5569 | Yes | ||
| 40 | CDK4 | 1422441_x_at | 1669 | 0.757 | 0.5613 | Yes | ||
| 41 | CENPE | 1435005_at 1439040_at | 1719 | 0.732 | 0.5655 | Yes | ||
| 42 | ORC1L | 1422663_at 1443172_at | 1723 | 0.732 | 0.5717 | Yes | ||
| 43 | CDC25C | 1422252_a_at | 1745 | 0.723 | 0.5771 | Yes | ||
| 44 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 1775 | 0.711 | 0.5820 | Yes | ||
| 45 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 1778 | 0.711 | 0.5881 | Yes | ||
| 46 | PSMC1 | 1416005_at | 1843 | 0.691 | 0.5912 | Yes | ||
| 47 | PSMA7 | 1423567_a_at 1423568_at | 1912 | 0.664 | 0.5939 | Yes | ||
| 48 | MCM2 | 1434079_s_at 1448777_at | 1953 | 0.651 | 0.5977 | Yes | ||
| 49 | PSMC4 | 1416290_a_at 1416291_at | 1988 | 0.642 | 0.6018 | Yes | ||
| 50 | PSMB10 | 1448632_at | 2021 | 0.629 | 0.6058 | Yes | ||
| 51 | NUP37 | 1423969_at | 2045 | 0.622 | 0.6102 | Yes | ||
| 52 | MAD2L1 | 1422460_at | 2085 | 0.611 | 0.6137 | Yes | ||
| 53 | PSMA4 | 1460339_at | 2170 | 0.587 | 0.6150 | Yes | ||
| 54 | TUBG1 | 1417144_at | 2211 | 0.577 | 0.6182 | Yes | ||
| 55 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 2279 | 0.560 | 0.6200 | Yes | ||
| 56 | PSMB5 | 1415676_a_at | 2348 | 0.542 | 0.6216 | Yes | ||
| 57 | BUB1 | 1424046_at 1438571_at | 2364 | 0.538 | 0.6256 | Yes | ||
| 58 | NUMA1 | 1423974_at 1423975_s_at 1441090_at 1444576_at 1444577_x_at | 2467 | 0.514 | 0.6254 | Yes | ||
| 59 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 2470 | 0.514 | 0.6298 | Yes | ||
| 60 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 2603 | 0.487 | 0.6280 | Yes | ||
| 61 | UBE2C | 1452954_at | 2626 | 0.483 | 0.6312 | Yes | ||
| 62 | PSMA2 | 1448206_at | 2629 | 0.483 | 0.6353 | Yes | ||
| 63 | CEP135 | 1444201_at | 2634 | 0.482 | 0.6394 | Yes | ||
| 64 | NUP160 | 1418530_at 1444892_at | 2779 | 0.455 | 0.6367 | No | ||
| 65 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 2905 | 0.433 | 0.6348 | No | ||
| 66 | PRKAR2B | 1430640_a_at 1438664_at 1456475_s_at | 3019 | 0.417 | 0.6332 | No | ||
| 67 | PSME3 | 1418078_at 1418079_at 1438509_at | 3225 | 0.385 | 0.6272 | No | ||
| 68 | CCNE1 | 1416492_at 1441910_x_at | 3304 | 0.375 | 0.6269 | No | ||
| 69 | PSMC3 | 1416282_at | 3317 | 0.373 | 0.6296 | No | ||
| 70 | PSMD3 | 1448479_at | 3472 | 0.355 | 0.6256 | No | ||
| 71 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 3497 | 0.351 | 0.6276 | No | ||
| 72 | SGOL2 | 1437370_at | 3632 | 0.338 | 0.6244 | No | ||
| 73 | PSMC5 | 1415740_at | 3784 | 0.326 | 0.6203 | No | ||
| 74 | SSNA1 | 1448643_at | 3969 | 0.311 | 0.6146 | No | ||
| 75 | DCTN3 | 1416247_at | 3998 | 0.308 | 0.6160 | No | ||
| 76 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 4174 | 0.296 | 0.6105 | No | ||
| 77 | FGFR1OP | 1428919_at | 4447 | 0.279 | 0.6005 | No | ||
| 78 | PSMD2 | 1415831_at | 4564 | 0.273 | 0.5975 | No | ||
| 79 | PSMD8 | 1423296_at | 4842 | 0.259 | 0.5871 | No | ||
| 80 | NUP107 | 1426751_s_at | 4920 | 0.256 | 0.5858 | No | ||
| 81 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 5504 | 0.231 | 0.5610 | No | ||
| 82 | CDK2 | 1416873_a_at 1447617_at | 5549 | 0.229 | 0.5610 | No | ||
| 83 | PLK1 | 1443408_at 1448191_at 1459616_at | 5819 | 0.219 | 0.5506 | No | ||
| 84 | CETN2 | 1418579_at 1438647_x_at | 6046 | 0.210 | 0.5420 | No | ||
| 85 | MCM4 | 1416214_at 1436708_x_at | 6049 | 0.210 | 0.5438 | No | ||
| 86 | PSMD5 | 1423234_at | 6087 | 0.209 | 0.5439 | No | ||
| 87 | CKAP5 | 1430647_at 1437349_at 1442271_at | 6204 | 0.204 | 0.5404 | No | ||
| 88 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 6944 | 0.180 | 0.5080 | No | ||
| 89 | PCNA | 1417947_at | 7044 | 0.177 | 0.5050 | No | ||
| 90 | PSMA3 | 1448442_a_at | 7498 | 0.164 | 0.4857 | No | ||
| 91 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 7608 | 0.160 | 0.4821 | No | ||
| 92 | ZW10 | 1424891_a_at 1431747_at 1445206_at | 8066 | 0.147 | 0.4624 | No | ||
| 93 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 8167 | 0.145 | 0.4590 | No | ||
| 94 | SMC1A | 1417830_at 1417831_at 1417832_at | 8283 | 0.142 | 0.4550 | No | ||
| 95 | CEP57 | 1428968_at 1452983_at | 8290 | 0.142 | 0.4560 | No | ||
| 96 | TUBB4 | 1423221_at | 8384 | 0.139 | 0.4529 | No | ||
| 97 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 8539 | 0.136 | 0.4470 | No | ||
| 98 | ORC5L | 1415830_at 1441601_at | 8593 | 0.134 | 0.4458 | No | ||
| 99 | CEP152 | 1427496_at | 8646 | 0.133 | 0.4446 | No | ||
| 100 | KNTC1 | 1435575_at | 9004 | 0.124 | 0.4293 | No | ||
| 101 | CUL1 | 1448174_at 1459516_at | 9352 | 0.115 | 0.4143 | No | ||
| 102 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 10055 | 0.098 | 0.3830 | No | ||
| 103 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 10479 | 0.089 | 0.3643 | No | ||
| 104 | PSMB4 | 1438984_x_at 1451205_at | 10735 | 0.083 | 0.3533 | No | ||
| 105 | PSMB2 | 1444377_at 1448262_at 1459641_at | 11183 | 0.073 | 0.3335 | No | ||
| 106 | KIF2A | 1432072_at 1450052_at 1450053_at 1452499_a_at 1454107_a_at | 11723 | 0.059 | 0.3092 | No | ||
| 107 | PRKACA | 1447720_x_at 1450519_a_at | 11849 | 0.056 | 0.3040 | No | ||
| 108 | DCTN2 | 1424461_at | 11901 | 0.055 | 0.3021 | No | ||
| 109 | CDC27 | 1426076_at 1433848_at 1433849_at 1454739_at | 11926 | 0.054 | 0.3015 | No | ||
| 110 | CENPC1 | 1420441_at | 12337 | 0.045 | 0.2831 | No | ||
| 111 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 12930 | 0.031 | 0.2562 | No | ||
| 112 | CEP78 | 1436352_at | 13231 | 0.023 | 0.2426 | No | ||
| 113 | DYNLL1 | 1448682_at | 13763 | 0.008 | 0.2183 | No | ||
| 114 | CEP110 | 1421004_at 1421005_at 1435779_at | 13771 | 0.008 | 0.2180 | No | ||
| 115 | RPA3 | 1448938_at | 14354 | -0.009 | 0.1914 | No | ||
| 116 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 14628 | -0.018 | 0.1790 | No | ||
| 117 | ACTR1A | 1415873_a_at 1458696_at 1459855_x_at | 14759 | -0.022 | 0.1733 | No | ||
| 118 | CCNA1 | 1449177_at | 14828 | -0.024 | 0.1703 | No | ||
| 119 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 15709 | -0.056 | 0.1304 | No | ||
| 120 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 15836 | -0.063 | 0.1252 | No | ||
| 121 | PLK4 | 1419838_s_at 1426580_at 1452115_a_at | 15985 | -0.070 | 0.1190 | No | ||
| 122 | RANBP2 | 1422621_at 1440104_at 1445883_at 1450690_at | 16137 | -0.077 | 0.1128 | No | ||
| 123 | XPO1 | 1418442_at 1418443_at 1448070_at | 16219 | -0.081 | 0.1098 | No | ||
| 124 | TK2 | 1426100_a_at | 16580 | -0.104 | 0.0941 | No | ||
| 125 | PSMD12 | 1448492_a_at | 16646 | -0.108 | 0.0921 | No | ||
| 126 | PSMC6 | 1417771_a_at | 16707 | -0.112 | 0.0903 | No | ||
| 127 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 17094 | -0.142 | 0.0738 | No | ||
| 128 | CCNH | 1418585_at | 17226 | -0.154 | 0.0692 | No | ||
| 129 | CDC16 | 1425554_a_at | 17441 | -0.175 | 0.0609 | No | ||
| 130 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 17505 | -0.181 | 0.0596 | No | ||
| 131 | MNAT1 | 1432177_a_at | 17605 | -0.191 | 0.0567 | No | ||
| 132 | OFD1 | 1427172_at | 17677 | -0.199 | 0.0552 | No | ||
| 133 | TAOK1 | 1424657_at 1424658_at 1426357_at 1426358_at 1444385_at 1455432_at 1456975_at 1459770_at | 17765 | -0.210 | 0.0530 | No | ||
| 134 | CDC25A | 1417131_at 1417132_at 1445995_at | 18425 | -0.297 | 0.0253 | No | ||
| 135 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 18543 | -0.312 | 0.0227 | No | ||
| 136 | SFI1 | 1428778_at | 18585 | -0.319 | 0.0236 | No | ||
| 137 | PSMD6 | 1423696_a_at 1423697_at | 18665 | -0.331 | 0.0229 | No | ||
| 138 | ORC2L | 1418225_at 1418226_at 1418227_at | 19158 | -0.431 | 0.0041 | No | ||
| 139 | RAD21 | 1416161_at 1416162_at 1455938_x_at | 19630 | -0.545 | -0.0128 | No | ||
| 140 | TUBB2C | 1423642_at 1438622_x_at 1439416_x_at 1456031_at 1456078_x_at 1456470_x_at | 19766 | -0.584 | -0.0139 | No | ||
| 141 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 19896 | -0.621 | -0.0144 | No | ||
| 142 | STAG2 | 1421849_at 1450396_at | 20247 | -0.758 | -0.0238 | No | ||
| 143 | PPP1CC | 1440594_at | 20391 | -0.821 | -0.0232 | No | ||
| 144 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 20420 | -0.832 | -0.0173 | No | ||
| 145 | DYNC1I2 | 1415841_at | 20515 | -0.877 | -0.0139 | No | ||
| 146 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 20627 | -0.935 | -0.0108 | No | ||
| 147 | SGOL1 | 1418919_at 1439510_at | 20771 | -1.047 | -0.0083 | No | ||
| 148 | CCNA2 | 1417910_at 1417911_at | 20779 | -1.050 | 0.0006 | No | ||
| 149 | UBE2D1 | 1424062_at | 20848 | -1.098 | 0.0071 | No | ||
| 150 | MCM3 | 1420029_at 1426653_at | 21483 | -1.983 | -0.0047 | No | ||
| 151 | CSNK1E | 1417175_at 1417176_at 1435373_at | 21762 | -2.918 | 0.0080 | No |