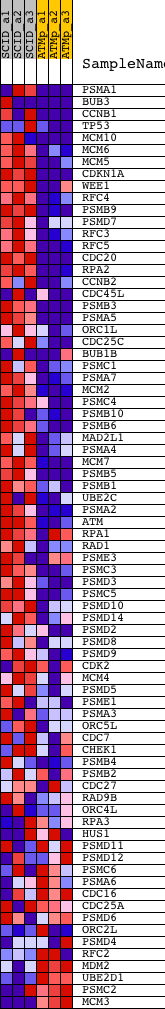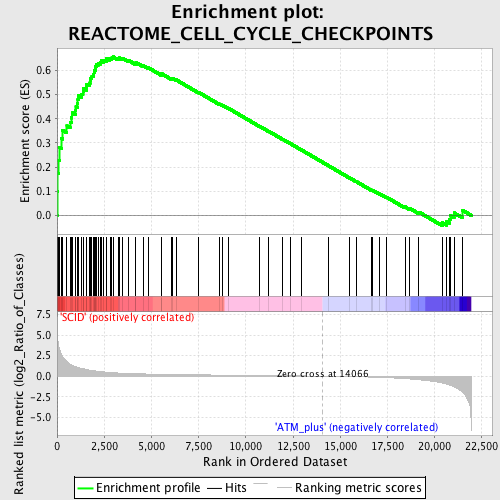
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | 0.6560747 |
| Normalized Enrichment Score (NES) | 1.906924 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0034561907 |
| FWER p-Value | 0.027 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 8 | 6.245 | 0.0979 | Yes | ||
| 2 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 24 | 4.996 | 0.1757 | Yes | ||
| 3 | CCNB1 | 1419943_s_at 1419944_at | 88 | 3.586 | 0.2292 | Yes | ||
| 4 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 111 | 3.368 | 0.2812 | Yes | ||
| 5 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 220 | 2.689 | 0.3186 | Yes | ||
| 6 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 280 | 2.372 | 0.3532 | Yes | ||
| 7 | MCM5 | 1415945_at 1436808_x_at | 521 | 1.778 | 0.3702 | Yes | ||
| 8 | CDKN1A | 1421679_a_at 1424638_at | 689 | 1.463 | 0.3855 | Yes | ||
| 9 | WEE1 | 1416773_at 1416774_at | 753 | 1.388 | 0.4045 | Yes | ||
| 10 | RFC4 | 1424321_at 1438161_s_at | 788 | 1.350 | 0.4242 | Yes | ||
| 11 | PSMB9 | 1450696_at | 978 | 1.164 | 0.4338 | Yes | ||
| 12 | PSMD7 | 1432820_at 1451056_at 1454368_at | 999 | 1.147 | 0.4509 | Yes | ||
| 13 | RFC3 | 1423700_at 1432538_a_at | 1086 | 1.072 | 0.4639 | Yes | ||
| 14 | RFC5 | 1452917_at | 1102 | 1.062 | 0.4799 | Yes | ||
| 15 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 1125 | 1.041 | 0.4953 | Yes | ||
| 16 | RPA2 | 1416433_at 1454011_a_at | 1307 | 0.930 | 0.5016 | Yes | ||
| 17 | CCNB2 | 1450920_at | 1372 | 0.898 | 0.5128 | Yes | ||
| 18 | CDC45L | 1416575_at 1457838_at | 1397 | 0.889 | 0.5257 | Yes | ||
| 19 | PSMB3 | 1417052_at | 1556 | 0.815 | 0.5313 | Yes | ||
| 20 | PSMA5 | 1424681_a_at 1434356_a_at | 1570 | 0.805 | 0.5433 | Yes | ||
| 21 | ORC1L | 1422663_at 1443172_at | 1723 | 0.732 | 0.5479 | Yes | ||
| 22 | CDC25C | 1422252_a_at | 1745 | 0.723 | 0.5583 | Yes | ||
| 23 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 1775 | 0.711 | 0.5682 | Yes | ||
| 24 | PSMC1 | 1416005_at | 1843 | 0.691 | 0.5760 | Yes | ||
| 25 | PSMA7 | 1423567_a_at 1423568_at | 1912 | 0.664 | 0.5833 | Yes | ||
| 26 | MCM2 | 1434079_s_at 1448777_at | 1953 | 0.651 | 0.5917 | Yes | ||
| 27 | PSMC4 | 1416290_a_at 1416291_at | 1988 | 0.642 | 0.6003 | Yes | ||
| 28 | PSMB10 | 1448632_at | 2021 | 0.629 | 0.6087 | Yes | ||
| 29 | PSMB6 | 1448822_at | 2044 | 0.622 | 0.6175 | Yes | ||
| 30 | MAD2L1 | 1422460_at | 2085 | 0.611 | 0.6252 | Yes | ||
| 31 | PSMA4 | 1460339_at | 2170 | 0.587 | 0.6306 | Yes | ||
| 32 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 2279 | 0.560 | 0.6345 | Yes | ||
| 33 | PSMB5 | 1415676_a_at | 2348 | 0.542 | 0.6399 | Yes | ||
| 34 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 2470 | 0.514 | 0.6424 | Yes | ||
| 35 | UBE2C | 1452954_at | 2626 | 0.483 | 0.6430 | Yes | ||
| 36 | PSMA2 | 1448206_at | 2629 | 0.483 | 0.6505 | Yes | ||
| 37 | ATM | 1421205_at 1428830_at | 2801 | 0.453 | 0.6498 | Yes | ||
| 38 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 2905 | 0.433 | 0.6518 | Yes | ||
| 39 | RAD1 | 1448414_at | 2960 | 0.426 | 0.6561 | Yes | ||
| 40 | PSME3 | 1418078_at 1418079_at 1438509_at | 3225 | 0.385 | 0.6500 | No | ||
| 41 | PSMC3 | 1416282_at | 3317 | 0.373 | 0.6518 | No | ||
| 42 | PSMD3 | 1448479_at | 3472 | 0.355 | 0.6503 | No | ||
| 43 | PSMC5 | 1415740_at | 3784 | 0.326 | 0.6412 | No | ||
| 44 | PSMD10 | 1436559_a_at | 4168 | 0.296 | 0.6283 | No | ||
| 45 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 4174 | 0.296 | 0.6328 | No | ||
| 46 | PSMD2 | 1415831_at | 4564 | 0.273 | 0.6193 | No | ||
| 47 | PSMD8 | 1423296_at | 4842 | 0.259 | 0.6107 | No | ||
| 48 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 5504 | 0.231 | 0.5841 | No | ||
| 49 | CDK2 | 1416873_a_at 1447617_at | 5549 | 0.229 | 0.5856 | No | ||
| 50 | MCM4 | 1416214_at 1436708_x_at | 6049 | 0.210 | 0.5661 | No | ||
| 51 | PSMD5 | 1423234_at | 6087 | 0.209 | 0.5677 | No | ||
| 52 | PSME1 | 1417056_at | 6301 | 0.201 | 0.5611 | No | ||
| 53 | PSMA3 | 1448442_a_at | 7498 | 0.164 | 0.5090 | No | ||
| 54 | ORC5L | 1415830_at 1441601_at | 8593 | 0.134 | 0.4611 | No | ||
| 55 | CDC7 | 1426002_a_at 1426021_a_at | 8770 | 0.130 | 0.4551 | No | ||
| 56 | CHEK1 | 1420031_at 1420032_at 1439208_at 1449708_s_at 1450677_at | 9076 | 0.122 | 0.4430 | No | ||
| 57 | PSMB4 | 1438984_x_at 1451205_at | 10735 | 0.083 | 0.3685 | No | ||
| 58 | PSMB2 | 1444377_at 1448262_at 1459641_at | 11183 | 0.073 | 0.3492 | No | ||
| 59 | CDC27 | 1426076_at 1433848_at 1433849_at 1454739_at | 11926 | 0.054 | 0.3161 | No | ||
| 60 | RAD9B | 1425800_at 1439955_at | 12374 | 0.044 | 0.2963 | No | ||
| 61 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 12930 | 0.031 | 0.2714 | No | ||
| 62 | RPA3 | 1448938_at | 14354 | -0.009 | 0.2065 | No | ||
| 63 | HUS1 | 1418308_at 1425366_a_at | 15503 | -0.049 | 0.1547 | No | ||
| 64 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 15836 | -0.063 | 0.1405 | No | ||
| 65 | PSMD12 | 1448492_a_at | 16646 | -0.108 | 0.1052 | No | ||
| 66 | PSMC6 | 1417771_a_at | 16707 | -0.112 | 0.1042 | No | ||
| 67 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 17094 | -0.142 | 0.0888 | No | ||
| 68 | CDC16 | 1425554_a_at | 17441 | -0.175 | 0.0757 | No | ||
| 69 | CDC25A | 1417131_at 1417132_at 1445995_at | 18425 | -0.297 | 0.0354 | No | ||
| 70 | PSMD6 | 1423696_a_at 1423697_at | 18665 | -0.331 | 0.0297 | No | ||
| 71 | ORC2L | 1418225_at 1418226_at 1418227_at | 19158 | -0.431 | 0.0140 | No | ||
| 72 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 20420 | -0.832 | -0.0306 | No | ||
| 73 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 20627 | -0.935 | -0.0253 | No | ||
| 74 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 20769 | -1.045 | -0.0153 | No | ||
| 75 | UBE2D1 | 1424062_at | 20848 | -1.098 | -0.0017 | No | ||
| 76 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21028 | -1.281 | 0.0103 | No | ||
| 77 | MCM3 | 1420029_at 1426653_at | 21483 | -1.983 | 0.0207 | No |