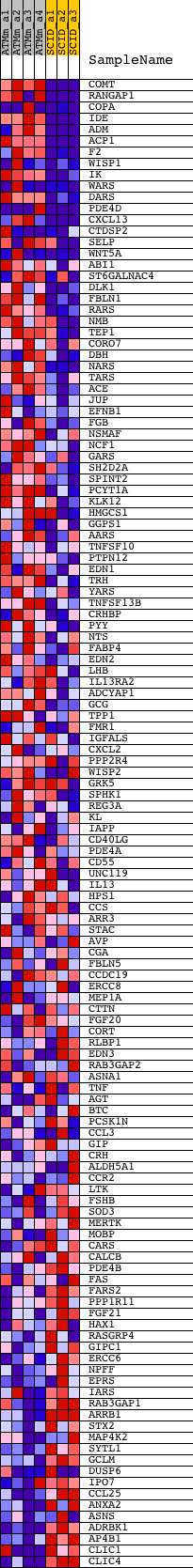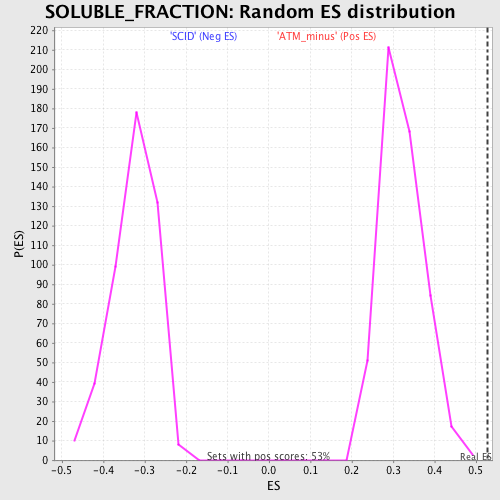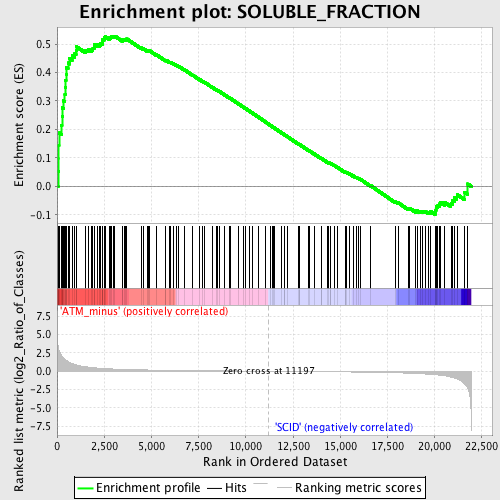
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | SOLUBLE_FRACTION |
| Enrichment Score (ES) | 0.52790546 |
| Normalized Enrichment Score (NES) | 1.6436827 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.55695677 |
| FWER p-Value | 0.989 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COMT | 1418701_at 1449183_at | 51 | 3.467 | 0.0523 | Yes | ||
| 2 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 93 | 2.929 | 0.0966 | Yes | ||
| 3 | COPA | 1415706_at 1437274_at | 95 | 2.920 | 0.1426 | Yes | ||
| 4 | IDE | 1423120_at 1423121_at 1435140_at 1441860_x_at 1453988_a_at | 100 | 2.873 | 0.1877 | Yes | ||
| 5 | ADM | 1416077_at 1447839_x_at | 237 | 2.110 | 0.2148 | Yes | ||
| 6 | ACP1 | 1422716_a_at 1422717_at 1450720_at 1450721_at | 259 | 2.027 | 0.2458 | Yes | ||
| 7 | F2 | 1418897_at | 277 | 1.972 | 0.2761 | Yes | ||
| 8 | WISP1 | 1448593_at 1448594_at | 348 | 1.744 | 0.3004 | Yes | ||
| 9 | IK | 1423791_at 1435988_x_at 1439271_x_at 1451113_a_at | 414 | 1.633 | 0.3231 | Yes | ||
| 10 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 426 | 1.610 | 0.3480 | Yes | ||
| 11 | DARS | 1423800_at | 454 | 1.556 | 0.3713 | Yes | ||
| 12 | PDE4D | 1443289_at 1458212_at 1459289_at 1459311_at | 472 | 1.517 | 0.3944 | Yes | ||
| 13 | CXCL13 | 1417851_at 1448859_at | 482 | 1.482 | 0.4174 | Yes | ||
| 14 | CTDSP2 | 1423660_at 1423661_s_at 1451075_s_at | 579 | 1.311 | 0.4337 | Yes | ||
| 15 | SELP | 1420558_at 1440173_x_at 1449906_at | 666 | 1.192 | 0.4485 | Yes | ||
| 16 | WNT5A | 1436791_at 1448818_at 1456976_at | 793 | 1.053 | 0.4593 | Yes | ||
| 17 | ABI1 | 1423177_a_at 1423178_at 1450890_a_at | 940 | 0.924 | 0.4672 | Yes | ||
| 18 | ST6GALNAC4 | 1418074_at 1418075_at | 1003 | 0.878 | 0.4782 | Yes | ||
| 19 | DLK1 | 1449939_s_at | 1019 | 0.867 | 0.4912 | Yes | ||
| 20 | FBLN1 | 1422540_at 1439688_at 1451119_a_at | 1510 | 0.613 | 0.4784 | Yes | ||
| 21 | RARS | 1416312_at | 1637 | 0.572 | 0.4816 | Yes | ||
| 22 | NMB | 1419405_at | 1843 | 0.510 | 0.4803 | Yes | ||
| 23 | TEP1 | 1418196_at | 1896 | 0.497 | 0.4857 | Yes | ||
| 24 | CORO7 | 1428147_at 1428148_s_at 1428149_at 1428150_at 1437146_x_at 1437524_x_at | 1961 | 0.481 | 0.4904 | Yes | ||
| 25 | DBH | 1447592_at 1450670_at 1459848_x_at | 1971 | 0.477 | 0.4975 | Yes | ||
| 26 | NARS | 1428666_at 1452866_at | 2121 | 0.439 | 0.4976 | Yes | ||
| 27 | TARS | 1460323_at | 2246 | 0.414 | 0.4984 | Yes | ||
| 28 | ACE | 1427034_at 1451911_a_at | 2313 | 0.401 | 0.5017 | Yes | ||
| 29 | JUP | 1426873_s_at | 2387 | 0.386 | 0.5044 | Yes | ||
| 30 | EFNB1 | 1418285_at 1418286_a_at 1451591_a_at | 2411 | 0.382 | 0.5094 | Yes | ||
| 31 | FGB | 1428079_at | 2419 | 0.381 | 0.5151 | Yes | ||
| 32 | NSMAF | 1416412_at | 2485 | 0.372 | 0.5180 | Yes | ||
| 33 | NCF1 | 1425609_at 1451767_at 1456772_at | 2513 | 0.366 | 0.5225 | Yes | ||
| 34 | GARS | 1423784_at | 2571 | 0.357 | 0.5255 | Yes | ||
| 35 | SH2D2A | 1449105_at | 2766 | 0.327 | 0.5218 | Yes | ||
| 36 | SPINT2 | 1438968_x_at 1451935_a_at | 2830 | 0.320 | 0.5239 | Yes | ||
| 37 | PCYT1A | 1421957_a_at 1424453_at 1438011_at 1450434_s_at | 2882 | 0.314 | 0.5266 | Yes | ||
| 38 | KLK12 | 1453495_at | 2992 | 0.300 | 0.5263 | Yes | ||
| 39 | HMGCS1 | 1433443_a_at 1433444_at 1433445_x_at 1433446_at 1441536_at | 3058 | 0.292 | 0.5279 | Yes | ||
| 40 | GGPS1 | 1419505_a_at 1419506_at 1419805_s_at 1429356_s_at | 3448 | 0.253 | 0.5140 | No | ||
| 41 | AARS | 1423685_at 1451083_s_at | 3466 | 0.251 | 0.5172 | No | ||
| 42 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 3591 | 0.240 | 0.5153 | No | ||
| 43 | PTPN12 | 1422045_a_at 1439705_at 1450478_a_at 1450479_x_at 1455105_at 1459459_at | 3645 | 0.237 | 0.5166 | No | ||
| 44 | EDN1 | 1451924_a_at | 3698 | 0.234 | 0.5179 | No | ||
| 45 | TRH | 1418756_at | 4444 | 0.185 | 0.4867 | No | ||
| 46 | YARS | 1460638_at | 4562 | 0.179 | 0.4841 | No | ||
| 47 | TNFSF13B | 1445251_at 1458047_at 1460255_at | 4804 | 0.167 | 0.4757 | No | ||
| 48 | CRHBP | 1436127_at | 4860 | 0.164 | 0.4758 | No | ||
| 49 | PYY | 1424865_at | 4871 | 0.164 | 0.4779 | No | ||
| 50 | NTS | 1422860_at | 5259 | 0.147 | 0.4625 | No | ||
| 51 | FABP4 | 1417023_a_at 1424155_at 1425809_at 1451263_a_at | 5742 | 0.130 | 0.4424 | No | ||
| 52 | EDN2 | 1449161_at | 5762 | 0.129 | 0.4436 | No | ||
| 53 | LHB | 1450795_at | 5971 | 0.122 | 0.4360 | No | ||
| 54 | IL13RA2 | 1422177_at | 5999 | 0.121 | 0.4366 | No | ||
| 55 | ADCYAP1 | 1423427_at 1441778_at | 6149 | 0.117 | 0.4316 | No | ||
| 56 | GCG | 1425952_a_at | 6329 | 0.111 | 0.4252 | No | ||
| 57 | TPP1 | 1434768_at 1448313_at | 6434 | 0.108 | 0.4221 | No | ||
| 58 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 6725 | 0.099 | 0.4104 | No | ||
| 59 | IGFALS | 1422826_at | 7172 | 0.087 | 0.3913 | No | ||
| 60 | CXCL2 | 1449984_at | 7561 | 0.077 | 0.3747 | No | ||
| 61 | PPP2R4 | 1439383_x_at 1439393_x_at 1448138_at | 7698 | 0.074 | 0.3697 | No | ||
| 62 | WISP2 | 1419015_at | 7822 | 0.071 | 0.3652 | No | ||
| 63 | GRK5 | 1446361_at 1446998_at 1449514_at | 8221 | 0.063 | 0.3479 | No | ||
| 64 | SPHK1 | 1451596_a_at | 8451 | 0.057 | 0.3383 | No | ||
| 65 | REG3A | 1449495_at | 8515 | 0.055 | 0.3363 | No | ||
| 66 | KL | 1423400_at 1451892_at | 8607 | 0.053 | 0.3329 | No | ||
| 67 | IAPP | 1423509_a_at 1423510_at 1437323_a_at 1456004_x_at | 8880 | 0.047 | 0.3212 | No | ||
| 68 | CD40LG | 1422283_at | 9135 | 0.041 | 0.3102 | No | ||
| 69 | PDE4A | 1421535_a_at 1445946_at 1457408_at | 9208 | 0.040 | 0.3075 | No | ||
| 70 | CD55 | 1418762_at 1443906_at 1460242_at | 9584 | 0.032 | 0.2908 | No | ||
| 71 | UNC119 | 1418123_at | 9866 | 0.026 | 0.2784 | No | ||
| 72 | IL13 | 1420802_at | 9962 | 0.024 | 0.2744 | No | ||
| 73 | HPS1 | 1443034_at 1448408_at 1459598_at 1459827_x_at | 10186 | 0.020 | 0.2645 | No | ||
| 74 | CCS | 1448615_at | 10346 | 0.017 | 0.2575 | No | ||
| 75 | ARR3 | 1425232_x_at 1441144_at 1450329_a_at | 10660 | 0.011 | 0.2433 | No | ||
| 76 | STAC | 1418957_at 1445344_at | 11012 | 0.004 | 0.2272 | No | ||
| 77 | AVP | 1450794_at | 11290 | -0.002 | 0.2146 | No | ||
| 78 | CGA | 1418549_at | 11432 | -0.005 | 0.2082 | No | ||
| 79 | FBLN5 | 1416164_at 1446377_at | 11438 | -0.005 | 0.2080 | No | ||
| 80 | CCDC19 | 1429930_at | 11497 | -0.007 | 0.2055 | No | ||
| 81 | ERCC8 | 1427194_a_at 1429439_at 1432249_a_at 1454176_at | 11870 | -0.014 | 0.1887 | No | ||
| 82 | MEP1A | 1447252_s_at 1450719_at | 12036 | -0.018 | 0.1814 | No | ||
| 83 | CTTN | 1421313_s_at 1421314_at 1421315_s_at 1423917_a_at 1433908_a_at | 12060 | -0.018 | 0.1806 | No | ||
| 84 | FGF20 | 1421677_at | 12189 | -0.021 | 0.1751 | No | ||
| 85 | CORT | 1449820_at | 12806 | -0.034 | 0.1474 | No | ||
| 86 | RLBP1 | 1418310_a_at | 12822 | -0.035 | 0.1472 | No | ||
| 87 | EDN3 | 1421136_at 1438696_at 1441923_s_at 1441924_x_at | 12852 | -0.036 | 0.1464 | No | ||
| 88 | RAB3GAP2 | 1436272_at 1454915_at | 13310 | -0.046 | 0.1262 | No | ||
| 89 | ASNA1 | 1418292_at | 13370 | -0.047 | 0.1242 | No | ||
| 90 | TNF | 1419607_at | 13611 | -0.053 | 0.1141 | No | ||
| 91 | AGT | 1423396_at | 13996 | -0.062 | 0.0974 | No | ||
| 92 | BTC | 1421161_at 1435541_at | 14014 | -0.062 | 0.0976 | No | ||
| 93 | PCSK1N | 1416965_at | 14311 | -0.070 | 0.0852 | No | ||
| 94 | CCL3 | 1419561_at | 14397 | -0.073 | 0.0824 | No | ||
| 95 | GIP | 1449908_at | 14484 | -0.075 | 0.0796 | No | ||
| 96 | CRH | 1457984_at | 14488 | -0.075 | 0.0807 | No | ||
| 97 | ALDH5A1 | 1453065_at | 14685 | -0.080 | 0.0730 | No | ||
| 98 | CCR2 | 1421186_at 1421187_at 1421188_at 1460067_at | 14827 | -0.084 | 0.0678 | No | ||
| 99 | LTK | 1460300_a_at | 15250 | -0.096 | 0.0500 | No | ||
| 100 | FSHB | 1450996_at | 15276 | -0.097 | 0.0504 | No | ||
| 101 | SOD3 | 1417633_at 1417634_at | 15321 | -0.098 | 0.0499 | No | ||
| 102 | MERTK | 1422869_at | 15476 | -0.102 | 0.0444 | No | ||
| 103 | MOBP | 1421010_at 1433785_at 1436263_at 1441308_at 1445656_at 1450088_a_at | 15682 | -0.109 | 0.0368 | No | ||
| 104 | CARS | 1427330_at 1452394_at | 15841 | -0.114 | 0.0313 | No | ||
| 105 | CALCB | 1422639_at | 15950 | -0.117 | 0.0282 | No | ||
| 106 | PDE4B | 1422473_at 1422474_at 1442700_at 1444731_at 1445449_at 1446577_at 1447237_at 1447718_at | 16048 | -0.121 | 0.0257 | No | ||
| 107 | FAS | 1460251_at | 16616 | -0.140 | 0.0019 | No | ||
| 108 | FARS2 | 1431354_a_at 1439406_x_at | 17903 | -0.208 | -0.0538 | No | ||
| 109 | PPP1R11 | 1417060_at 1448565_at | 18059 | -0.219 | -0.0575 | No | ||
| 110 | FGF21 | 1422916_at | 18601 | -0.264 | -0.0781 | No | ||
| 111 | HAX1 | 1426187_a_at | 18655 | -0.269 | -0.0763 | No | ||
| 112 | RASGRP4 | 1425380_at | 18978 | -0.302 | -0.0863 | No | ||
| 113 | GIPC1 | 1422763_at | 19080 | -0.314 | -0.0860 | No | ||
| 114 | ERCC6 | 1441595_at 1442604_at | 19248 | -0.339 | -0.0883 | No | ||
| 115 | NPFF | 1420606_at | 19348 | -0.352 | -0.0873 | No | ||
| 116 | EPRS | 1430370_at | 19490 | -0.373 | -0.0879 | No | ||
| 117 | IARS | 1426705_s_at 1452154_at | 19687 | -0.404 | -0.0905 | No | ||
| 118 | RAB3GAP1 | 1455922_at | 19784 | -0.424 | -0.0882 | No | ||
| 119 | ARRB1 | 1460444_at | 20020 | -0.473 | -0.0916 | No | ||
| 120 | STX2 | 1418164_at | 20054 | -0.483 | -0.0854 | No | ||
| 121 | MAP4K2 | 1428297_at 1434833_at 1450244_a_at | 20070 | -0.486 | -0.0785 | No | ||
| 122 | SYTL1 | 1450240_a_at | 20074 | -0.487 | -0.0709 | No | ||
| 123 | GCLM | 1418627_at | 20168 | -0.513 | -0.0671 | No | ||
| 124 | DUSP6 | 1415834_at | 20242 | -0.535 | -0.0620 | No | ||
| 125 | IPO7 | 1442511_at 1454955_at 1458165_at | 20314 | -0.555 | -0.0565 | No | ||
| 126 | CCL25 | 1418777_at 1458277_at | 20524 | -0.625 | -0.0562 | No | ||
| 127 | ANXA2 | 1419091_a_at 1437691_at 1437692_x_at | 20862 | -0.793 | -0.0592 | No | ||
| 128 | ASNS | 1433966_x_at 1451095_at | 20945 | -0.850 | -0.0495 | No | ||
| 129 | ADRBK1 | 1426249_at 1451992_at | 21052 | -0.933 | -0.0397 | No | ||
| 130 | AP4B1 | 1416631_at 1421726_at | 21179 | -1.061 | -0.0287 | No | ||
| 131 | CLIC1 | 1416656_at 1458665_at | 21564 | -1.653 | -0.0203 | No | ||
| 132 | CLIC4 | 1423392_at 1423393_at 1438606_a_at | 21756 | -2.366 | 0.0083 | No |