Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
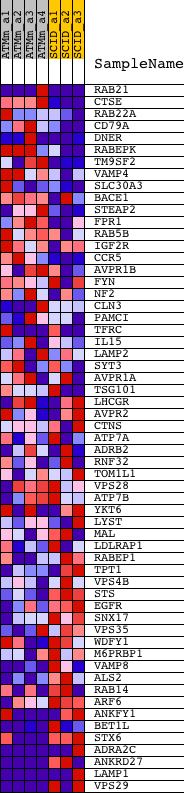
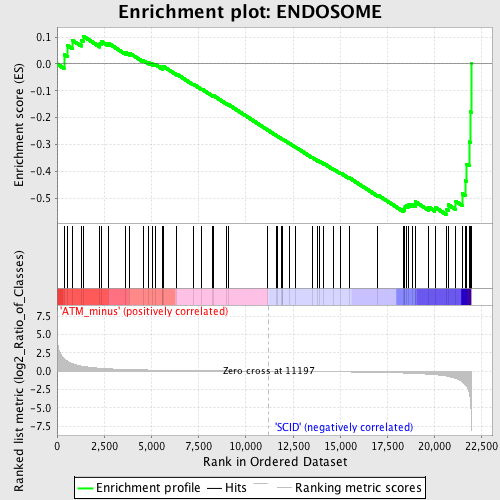
| GeneSet | ENDOSOME |
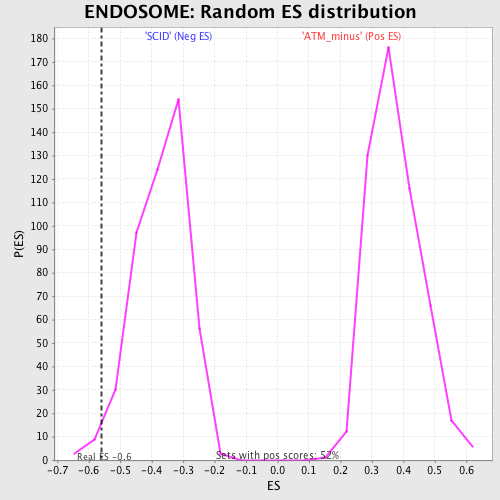
| Enrichment Score (ES) | -0.5608646 |
| Normalized Enrichment Score (NES) | -1.5191603 |
| Nominal p-value | 0.016806724 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RAB21 | 1437741_at 1437742_at 1447497_at | 367 | 1.708 | 0.0355 | No | ||
| 2 | CTSE | 1418989_at | 561 | 1.344 | 0.0678 | No | ||
| 3 | RAB22A | 1424503_at 1424504_at 1451379_at 1454988_s_at | 813 | 1.028 | 0.0879 | No | ||
| 4 | CD79A | 1418830_at | 1274 | 0.702 | 0.0883 | No | ||
| 5 | DNER | 1423671_at 1456379_x_at | 1378 | 0.658 | 0.1038 | No | ||
| 6 | RABEPK | 1433041_at 1448046_at 1451487_at 1454449_at | 2269 | 0.409 | 0.0756 | No | ||
| 7 | TM9SF2 | 1423295_at 1455875_x_at 1456742_x_at 1460331_at | 2369 | 0.389 | 0.0830 | No | ||
| 8 | VAMP4 | 1422895_at 1422896_at 1434796_at | 2719 | 0.335 | 0.0773 | No | ||
| 9 | SLC30A3 | 1460654_at | 3629 | 0.238 | 0.0430 | No | ||
| 10 | BACE1 | 1421824_at 1421825_at 1436956_at 1450384_at 1455826_a_at | 3846 | 0.223 | 0.0400 | No | ||
| 11 | STEAP2 | 1428636_at 1438773_at 1442359_at 1444290_at 1446071_at | 4550 | 0.179 | 0.0133 | No | ||
| 12 | FPR1 | 1450808_at | 4848 | 0.165 | 0.0048 | No | ||
| 13 | RAB5B | 1422119_at 1437446_at | 5055 | 0.156 | 0.0002 | No | ||
| 14 | IGF2R | 1424111_at 1424112_at 1440979_at 1445966_at 1457356_at | 5214 | 0.149 | -0.0025 | No | ||
| 15 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 5577 | 0.135 | -0.0149 | No | ||
| 16 | AVPR1B | 1422204_at | 5585 | 0.135 | -0.0111 | No | ||
| 17 | FYN | 1417558_at 1441647_at 1448765_at | 5640 | 0.133 | -0.0095 | No | ||
| 18 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 6347 | 0.111 | -0.0384 | No | ||
| 19 | CLN3 | 1417551_at 1457789_at | 7205 | 0.087 | -0.0749 | No | ||
| 20 | PAMCI | 1427942_at | 7661 | 0.075 | -0.0934 | No | ||
| 21 | TFRC | 1422966_a_at 1422967_a_at 1452661_at AFFX-TransRecMur/X57349_3_at AFFX-TransRecMur/X57349_5_at AFFX-TransRecMur/X57349_M_at | 8244 | 0.062 | -0.1181 | No | ||
| 22 | IL15 | 1418219_at | 8258 | 0.062 | -0.1168 | No | ||
| 23 | LAMP2 | 1416343_a_at 1416344_at 1428094_at 1434503_s_at | 8978 | 0.045 | -0.1483 | No | ||
| 24 | SYT3 | 1417708_at | 9090 | 0.042 | -0.1520 | No | ||
| 25 | AVPR1A | 1418603_at 1418604_at | 11159 | 0.001 | -0.2465 | No | ||
| 26 | TSG101 | 1417004_at | 11605 | -0.009 | -0.2666 | No | ||
| 27 | LHCGR | 1450192_at | 11654 | -0.010 | -0.2685 | No | ||
| 28 | AVPR2 | 1450277_at | 11883 | -0.015 | -0.2785 | No | ||
| 29 | CTNS | 1416274_at 1456450_at | 11953 | -0.016 | -0.2811 | No | ||
| 30 | ATP7A | 1418774_a_at 1436921_at | 11959 | -0.016 | -0.2809 | No | ||
| 31 | ADRB2 | 1437302_at | 12302 | -0.023 | -0.2958 | No | ||
| 32 | RNF32 | 1431365_at 1432156_a_at 1453666_at | 12622 | -0.030 | -0.3094 | No | ||
| 33 | TOM1L1 | 1439337_at 1451117_a_at | 13505 | -0.050 | -0.3482 | No | ||
| 34 | VPS28 | 1448447_at | 13812 | -0.057 | -0.3604 | No | ||
| 35 | ATP7B | 1421563_at 1446004_at 1450283_at | 13920 | -0.060 | -0.3635 | No | ||
| 36 | YKT6 | 1448464_at 1460191_at | 14130 | -0.065 | -0.3710 | No | ||
| 37 | LYST | 1421384_at 1425973_at 1434674_at | 14654 | -0.079 | -0.3925 | No | ||
| 38 | MAL | 1417275_at 1432558_a_at | 15035 | -0.089 | -0.4071 | No | ||
| 39 | LDLRAP1 | 1424378_at 1457162_at | 15459 | -0.102 | -0.4234 | No | ||
| 40 | RABEP1 | 1421305_x_at 1426023_a_at 1427448_at 1443856_at 1444930_at 1459494_at | 16984 | -0.155 | -0.4883 | No | ||
| 41 | TPT1 | 1416642_a_at 1416643_at | 18341 | -0.240 | -0.5429 | Yes | ||
| 42 | VPS4B | 1417007_a_at 1457067_at 1460104_at | 18390 | -0.245 | -0.5376 | Yes | ||
| 43 | STS | 1421500_at | 18424 | -0.248 | -0.5315 | Yes | ||
| 44 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 18510 | -0.256 | -0.5276 | Yes | ||
| 45 | SNX17 | 1455955_s_at | 18633 | -0.268 | -0.5249 | Yes | ||
| 46 | VPS35 | 1415783_at 1415784_at | 18800 | -0.283 | -0.5239 | Yes | ||
| 47 | WDFY1 | 1424749_at 1435588_at 1437358_at 1447543_at 1453426_a_at | 18966 | -0.300 | -0.5222 | Yes | ||
| 48 | M6PRBP1 | 1416424_at | 18971 | -0.301 | -0.5132 | Yes | ||
| 49 | VAMP8 | 1420624_a_at | 19686 | -0.404 | -0.5335 | Yes | ||
| 50 | ALS2 | 1417783_at 1417784_at 1431608_at 1446183_at | 20014 | -0.472 | -0.5339 | Yes | ||
| 51 | RAB14 | 1415686_at 1419243_at 1419244_a_at 1419245_at 1419246_s_at 1439687_at 1441992_at | 20604 | -0.653 | -0.5409 | Yes | ||
| 52 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 20713 | -0.708 | -0.5241 | Yes | ||
| 53 | ANKFY1 | 1417685_at 1443654_at | 21122 | -1.001 | -0.5121 | Yes | ||
| 54 | BET1L | 1422980_a_at 1426372_a_at 1430549_at 1453570_x_at | 21452 | -1.436 | -0.4832 | Yes | ||
| 55 | STX6 | 1423038_at 1431646_a_at 1440585_at 1441133_at 1450844_at 1458920_at 1460004_x_at | 21627 | -1.836 | -0.4349 | Yes | ||
| 56 | ADRA2C | 1422335_at 1435296_at 1439762_x_at 1457328_at | 21700 | -2.096 | -0.3741 | Yes | ||
| 57 | ANKRD27 | 1425112_at 1454717_at | 21834 | -2.927 | -0.2905 | Yes | ||
| 58 | LAMP1 | 1415880_a_at 1460096_at | 21881 | -3.769 | -0.1772 | Yes | ||
| 59 | VPS29 | 1417659_at 1417660_s_at 1440641_at 1455302_at | 21929 | -5.869 | 0.0003 | Yes |