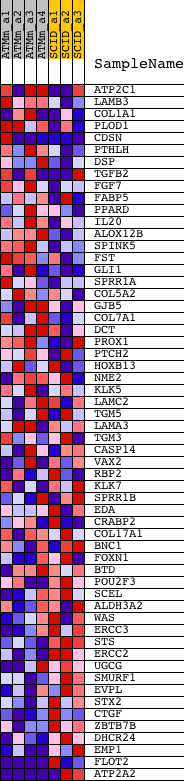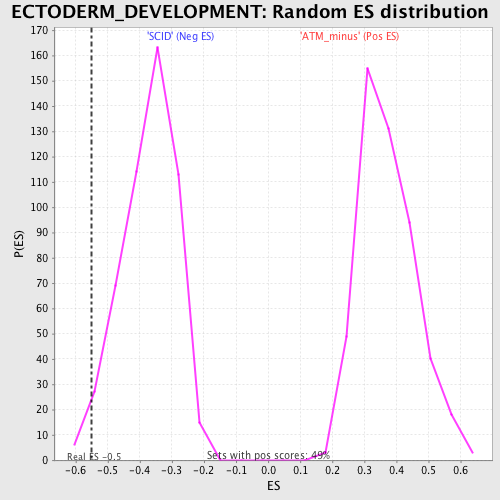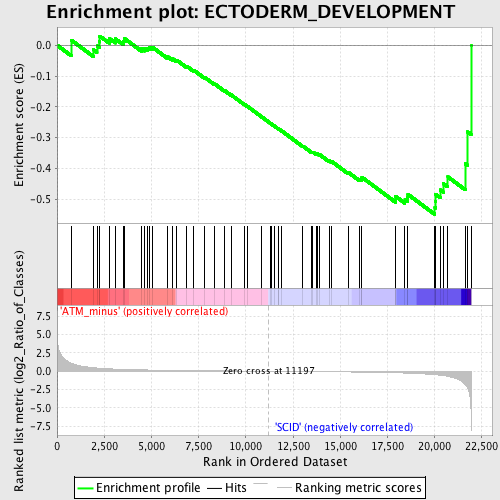
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | ECTODERM_DEVELOPMENT |
| Enrichment Score (ES) | -0.54988015 |
| Normalized Enrichment Score (NES) | -1.4783242 |
| Nominal p-value | 0.015779093 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 747 | 1.092 | 0.0168 | No | ||
| 2 | LAMB3 | 1417812_a_at | 1934 | 0.487 | -0.0147 | No | ||
| 3 | COL1A1 | 1423669_at 1455494_at | 2113 | 0.441 | -0.0023 | No | ||
| 4 | PLOD1 | 1416289_at 1445893_at | 2241 | 0.415 | 0.0113 | No | ||
| 5 | CDSN | 1435191_at 1444607_at | 2261 | 0.411 | 0.0296 | No | ||
| 6 | PTHLH | 1422324_a_at 1427527_a_at | 2759 | 0.328 | 0.0222 | No | ||
| 7 | DSP | 1427610_at 1435493_at 1435494_s_at | 3088 | 0.289 | 0.0206 | No | ||
| 8 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 3499 | 0.247 | 0.0134 | No | ||
| 9 | FGF7 | 1422243_at 1438405_at | 3564 | 0.243 | 0.0218 | No | ||
| 10 | FABP5 | 1416022_at | 4484 | 0.183 | -0.0117 | No | ||
| 11 | PPARD | 1425703_at 1439797_at | 4642 | 0.174 | -0.0107 | No | ||
| 12 | IL20 | 1421608_at | 4785 | 0.168 | -0.0094 | No | ||
| 13 | ALOX12B | 1418266_at | 4892 | 0.163 | -0.0066 | No | ||
| 14 | SPINK5 | 1430567_at | 5058 | 0.155 | -0.0069 | No | ||
| 15 | FST | 1421365_at 1434458_at | 5824 | 0.127 | -0.0360 | No | ||
| 16 | GLI1 | 1449058_at | 6101 | 0.118 | -0.0431 | No | ||
| 17 | SPRR1A | 1449133_at | 6326 | 0.111 | -0.0481 | No | ||
| 18 | COL5A2 | 1422437_at 1450625_at | 6850 | 0.096 | -0.0676 | No | ||
| 19 | GJB5 | 1449204_at | 7225 | 0.086 | -0.0806 | No | ||
| 20 | COL7A1 | 1419613_at | 7805 | 0.071 | -0.1038 | No | ||
| 21 | DCT | 1418028_at | 8348 | 0.059 | -0.1258 | No | ||
| 22 | PROX1 | 1421336_at 1437894_at 1457432_at | 8878 | 0.047 | -0.1477 | No | ||
| 23 | PTCH2 | 1422655_at | 9211 | 0.040 | -0.1611 | No | ||
| 24 | HOXB13 | 1419576_at | 9937 | 0.025 | -0.1931 | No | ||
| 25 | NME2 | 1448808_a_at | 10097 | 0.022 | -0.1993 | No | ||
| 26 | KLK5 | 1429230_at | 10849 | 0.007 | -0.2333 | No | ||
| 27 | LAMC2 | 1421279_at 1452505_at | 11322 | -0.003 | -0.2547 | No | ||
| 28 | TGM5 | 1430142_at | 11374 | -0.004 | -0.2569 | No | ||
| 29 | LAMA3 | 1444860_at | 11532 | -0.007 | -0.2637 | No | ||
| 30 | TGM3 | 1421355_at 1440150_at | 11699 | -0.011 | -0.2708 | No | ||
| 31 | CASP14 | 1418748_at 1443787_x_at | 11722 | -0.012 | -0.2713 | No | ||
| 32 | VAX2 | 1445218_at 1450303_at | 11889 | -0.015 | -0.2782 | No | ||
| 33 | RBP2 | 1422846_at | 13021 | -0.039 | -0.3280 | No | ||
| 34 | KLK7 | 1423542_at | 13490 | -0.050 | -0.3471 | No | ||
| 35 | SPRR1B | 1422672_at | 13550 | -0.051 | -0.3474 | No | ||
| 36 | EDA | 1419597_at 1449524_at 1451877_at 1451925_at | 13717 | -0.055 | -0.3524 | No | ||
| 37 | CRABP2 | 1451191_at | 13771 | -0.056 | -0.3522 | No | ||
| 38 | COL17A1 | 1418799_a_at | 13884 | -0.059 | -0.3546 | No | ||
| 39 | BNC1 | 1424890_at | 14407 | -0.073 | -0.3750 | No | ||
| 40 | FOXN1 | 1450508_at 1456815_at | 14520 | -0.076 | -0.3766 | No | ||
| 41 | BTD | 1417987_at | 15427 | -0.101 | -0.4133 | No | ||
| 42 | POU2F3 | 1450507_at | 16023 | -0.120 | -0.4349 | No | ||
| 43 | SCEL | 1422837_at 1459426_at | 16125 | -0.123 | -0.4338 | No | ||
| 44 | ALDH3A2 | 1415776_at | 16142 | -0.123 | -0.4288 | No | ||
| 45 | WAS | 1419631_at | 17917 | -0.209 | -0.5001 | No | ||
| 46 | ERCC3 | 1448497_at | 17921 | -0.209 | -0.4905 | No | ||
| 47 | STS | 1421500_at | 18424 | -0.248 | -0.5019 | No | ||
| 48 | ERCC2 | 1460727_at | 18555 | -0.260 | -0.4957 | No | ||
| 49 | UGCG | 1421268_at 1421269_at 1435133_at 1439863_at | 18580 | -0.263 | -0.4845 | No | ||
| 50 | SMURF1 | 1443393_at | 20011 | -0.472 | -0.5279 | Yes | ||
| 51 | EVPL | 1419133_at | 20038 | -0.479 | -0.5068 | Yes | ||
| 52 | STX2 | 1418164_at | 20054 | -0.483 | -0.4849 | Yes | ||
| 53 | CTGF | 1416953_at | 20292 | -0.550 | -0.4701 | Yes | ||
| 54 | ZBTB7B | 1420663_at 1455071_at | 20466 | -0.605 | -0.4498 | Yes | ||
| 55 | DHCR24 | 1418129_at 1451895_a_at | 20699 | -0.699 | -0.4278 | Yes | ||
| 56 | EMP1 | 1416529_at 1459171_at | 21618 | -1.820 | -0.3849 | Yes | ||
| 57 | FLOT2 | 1417544_a_at 1436666_at 1438164_x_at | 21750 | -2.342 | -0.2816 | Yes | ||
| 58 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 21930 | -6.219 | 0.0003 | Yes |