Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
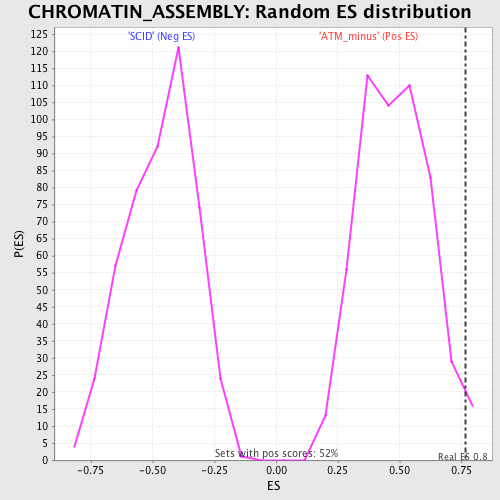
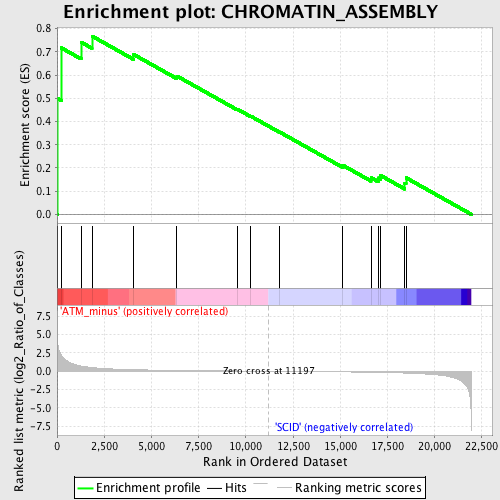
| GeneSet | CHROMATIN_ASSEMBLY |
| Enrichment Score (ES) | 0.7680404 |
| Normalized Enrichment Score (NES) | 1.5946139 |
| Nominal p-value | 0.022900764 |
| FDR q-value | 0.6277463 |
| FWER p-Value | 1.0 |

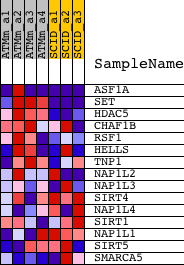
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ASF1A | 1423511_at 1459882_at | 14 | 4.863 | 0.5007 | Yes | ||
| 2 | SET | 1426853_at 1426854_a_at | 219 | 2.193 | 0.7175 | Yes | ||
| 3 | HDAC5 | 1415743_at | 1273 | 0.702 | 0.7418 | Yes | ||
| 4 | CHAF1B | 1423877_at 1431275_at | 1849 | 0.508 | 0.7680 | Yes | ||
| 5 | RSF1 | 1438735_at | 4041 | 0.208 | 0.6896 | No | ||
| 6 | HELLS | 1417541_at 1430139_at 1453361_at | 6346 | 0.111 | 0.5959 | No | ||
| 7 | TNP1 | 1415924_at 1438632_x_at | 9544 | 0.033 | 0.4535 | No | ||
| 8 | NAP1L2 | 1418046_at | 10262 | 0.019 | 0.4227 | No | ||
| 9 | NAP1L3 | 1418500_at | 11777 | -0.013 | 0.3549 | No | ||
| 10 | SIRT4 | 1426847_at | 15115 | -0.092 | 0.2122 | No | ||
| 11 | NAP1L4 | 1448476_at | 16625 | -0.141 | 0.1578 | No | ||
| 12 | SIRT1 | 1418640_at 1458538_at | 17005 | -0.156 | 0.1567 | No | ||
| 13 | NAP1L1 | 1420476_a_at 1420477_at 1420478_at 1420479_a_at 1429227_x_at 1437945_x_at 1440263_at 1452778_x_at | 17113 | -0.161 | 0.1684 | No | ||
| 14 | SIRT5 | 1428915_at 1428916_s_at 1432105_at | 18416 | -0.247 | 0.1344 | No | ||
| 15 | SMARCA5 | 1440048_at | 18481 | -0.253 | 0.1576 | No |