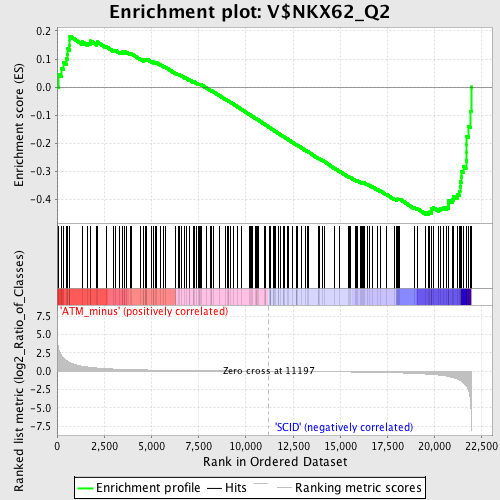
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | V$NKX62_Q2 |
| Enrichment Score (ES) | -0.45406854 |
| Normalized Enrichment Score (NES) | -1.4272428 |
| Nominal p-value | 0.008032128 |
| FDR q-value | 0.3602539 |
| FWER p-Value | 0.994 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | XPO1 | 1418442_at 1418443_at 1448070_at | 57 | 3.357 | 0.0452 | No | ||
| 2 | SERTAD4 | 1454877_at | 245 | 2.089 | 0.0664 | No | ||
| 3 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 326 | 1.803 | 0.0884 | No | ||
| 4 | DACH1 | 1420694_a_at 1420695_at 1433743_at 1447174_at 1459381_at | 506 | 1.433 | 0.1006 | No | ||
| 5 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 555 | 1.348 | 0.1176 | No | ||
| 6 | RAB6A | 1447776_x_at 1448304_a_at 1448305_at | 569 | 1.331 | 0.1360 | No | ||
| 7 | CYFIP2 | 1428347_at 1442167_at 1444351_at 1449273_at | 668 | 1.191 | 0.1485 | No | ||
| 8 | AIM1L | 1437813_at | 677 | 1.176 | 0.1649 | No | ||
| 9 | CYCS | 1422484_at 1445484_at | 679 | 1.174 | 0.1816 | No | ||
| 10 | IER3 | 1419647_a_at | 1328 | 0.678 | 0.1614 | No | ||
| 11 | FARP1 | 1445200_at 1452280_at 1459799_at | 1621 | 0.577 | 0.1563 | No | ||
| 12 | ZFPM2 | 1419012_at 1444803_at 1447020_at 1449314_at | 1754 | 0.538 | 0.1579 | No | ||
| 13 | STX7 | 1418436_at | 1779 | 0.532 | 0.1643 | No | ||
| 14 | AP1G1 | 1423388_at 1434081_at 1460658_at 1460712_s_at | 2098 | 0.444 | 0.1560 | No | ||
| 15 | PHF5A | 1424170_at | 2129 | 0.438 | 0.1609 | No | ||
| 16 | BCL9L | 1419180_at 1438806_at 1443700_at 1444855_at | 2594 | 0.353 | 0.1446 | No | ||
| 17 | ETV3 | 1418635_at 1418636_at 1445997_at 1456358_at | 3000 | 0.299 | 0.1303 | No | ||
| 18 | CYLD | 1429617_at 1429618_at 1445213_at | 3114 | 0.286 | 0.1292 | No | ||
| 19 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 3297 | 0.267 | 0.1246 | No | ||
| 20 | SSH2 | 1456153_at | 3439 | 0.253 | 0.1217 | No | ||
| 21 | ITGA7 | 1418393_a_at | 3440 | 0.253 | 0.1253 | No | ||
| 22 | NEUROD2 | 1444362_at | 3570 | 0.242 | 0.1229 | No | ||
| 23 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 3591 | 0.240 | 0.1254 | No | ||
| 24 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 3689 | 0.234 | 0.1242 | No | ||
| 25 | MAB21L1 | 1421369_a_at 1424679_at 1440860_at 1457290_at | 3891 | 0.219 | 0.1181 | No | ||
| 26 | ATOH1 | 1449822_at | 3918 | 0.217 | 0.1200 | No | ||
| 27 | PDZRN4 | 1456512_at | 4417 | 0.186 | 0.0998 | No | ||
| 28 | PRKAA1 | 1437539_at | 4579 | 0.178 | 0.0949 | No | ||
| 29 | NPAS2 | 1421036_at 1421037_at | 4594 | 0.177 | 0.0968 | No | ||
| 30 | IRX4 | 1419539_at | 4663 | 0.174 | 0.0962 | No | ||
| 31 | BMX | 1422084_at | 4691 | 0.172 | 0.0974 | No | ||
| 32 | FOXB1 | 1420705_at | 4709 | 0.171 | 0.0990 | No | ||
| 33 | BACE2 | 1416673_at 1435581_at 1437846_x_at 1438645_x_at | 4747 | 0.170 | 0.0998 | No | ||
| 34 | CACNA2D3 | 1419225_at | 5020 | 0.157 | 0.0895 | No | ||
| 35 | BAMBI | 1423753_at | 5109 | 0.153 | 0.0876 | No | ||
| 36 | SLIT3 | 1427086_at 1452296_at | 5117 | 0.153 | 0.0895 | No | ||
| 37 | MITF | 1422025_at 1455214_at | 5203 | 0.150 | 0.0877 | No | ||
| 38 | SOX15 | 1452017_at | 5276 | 0.147 | 0.0865 | No | ||
| 39 | PRIMA1 | 1425304_s_at | 5456 | 0.140 | 0.0803 | No | ||
| 40 | ARTN | 1432032_a_at | 5651 | 0.133 | 0.0733 | No | ||
| 41 | FABP4 | 1417023_a_at 1424155_at 1425809_at 1451263_a_at | 5742 | 0.130 | 0.0710 | No | ||
| 42 | ZIC4 | 1421539_at 1456417_at | 6275 | 0.113 | 0.0481 | No | ||
| 43 | ADAM11 | 1450248_at | 6282 | 0.112 | 0.0495 | No | ||
| 44 | IRX6 | 1427383_at | 6421 | 0.109 | 0.0447 | No | ||
| 45 | GREM1 | 1425357_a_at | 6483 | 0.106 | 0.0434 | No | ||
| 46 | KCNS1 | 1421518_at | 6578 | 0.104 | 0.0405 | No | ||
| 47 | PTHR1 | 1417092_at | 6772 | 0.098 | 0.0331 | No | ||
| 48 | NFIX | 1423493_a_at 1436363_a_at 1436364_x_at 1451443_at 1459909_at | 6837 | 0.096 | 0.0315 | No | ||
| 49 | GSH1 | 1450002_at | 7018 | 0.092 | 0.0245 | No | ||
| 50 | A2BP1 | 1418314_a_at 1438217_at 1447735_x_at 1455358_at 1456921_at | 7029 | 0.091 | 0.0254 | No | ||
| 51 | ERG | 1425370_a_at 1440244_at | 7200 | 0.087 | 0.0188 | No | ||
| 52 | LRRTM4 | 1437214_at 1455937_at | 7231 | 0.086 | 0.0187 | No | ||
| 53 | WNT3A | 1422093_at 1446764_at | 7253 | 0.085 | 0.0189 | No | ||
| 54 | RHOBTB1 | 1429206_at 1429656_at | 7383 | 0.082 | 0.0142 | No | ||
| 55 | KCNN3 | 1421632_at 1459308_at | 7487 | 0.079 | 0.0106 | No | ||
| 56 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 7542 | 0.078 | 0.0092 | No | ||
| 57 | LRFN5 | 1441502_at 1454687_at | 7544 | 0.078 | 0.0102 | No | ||
| 58 | IGSF4D | 1435145_at 1435146_s_at 1435147_x_at 1436743_at 1440412_at 1444664_at 1459761_x_at | 7591 | 0.077 | 0.0092 | No | ||
| 59 | PAMCI | 1427942_at | 7661 | 0.075 | 0.0071 | No | ||
| 60 | SLC37A4 | 1417042_at | 7896 | 0.070 | -0.0026 | No | ||
| 61 | PDGFRA | 1421916_at 1421917_at 1438946_at | 8143 | 0.064 | -0.0130 | No | ||
| 62 | PGR | 1421444_at 1439699_at 1455772_at | 8172 | 0.064 | -0.0134 | No | ||
| 63 | SOX14 | 1427609_at | 8174 | 0.064 | -0.0125 | No | ||
| 64 | FLOT1 | 1448559_at | 8277 | 0.061 | -0.0163 | No | ||
| 65 | FLRT3 | 1429310_at 1453102_at | 8602 | 0.053 | -0.0305 | No | ||
| 66 | TCEAL1 | 1424634_at | 8906 | 0.046 | -0.0437 | No | ||
| 67 | ANGPT1 | 1421441_at 1438936_s_at 1438937_x_at 1439066_at 1446656_at 1450717_at 1457381_at | 9034 | 0.043 | -0.0489 | No | ||
| 68 | PTF1A | 1419424_at | 9072 | 0.043 | -0.0500 | No | ||
| 69 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 9075 | 0.043 | -0.0495 | No | ||
| 70 | SCRT2 | 1440930_a_at 1440931_at 1446986_at 1457310_x_at | 9177 | 0.040 | -0.0536 | No | ||
| 71 | STMN2 | 1423280_at 1423281_at | 9350 | 0.037 | -0.0610 | No | ||
| 72 | TBX5 | 1425694_at 1425695_at | 9578 | 0.032 | -0.0709 | No | ||
| 73 | C1QTNF3 | 1422606_at | 9752 | 0.028 | -0.0785 | No | ||
| 74 | HOXA11 | 1420414_at | 10190 | 0.020 | -0.0983 | No | ||
| 75 | IRAK1 | 1448668_a_at 1460649_at | 10222 | 0.020 | -0.0994 | No | ||
| 76 | SLN | 1420883_at 1420884_at | 10292 | 0.018 | -0.1023 | No | ||
| 77 | TWIST1 | 1418733_at | 10365 | 0.017 | -0.1054 | No | ||
| 78 | PELI2 | 1419006_s_at 1436966_at 1437181_at | 10518 | 0.014 | -0.1122 | No | ||
| 79 | FLRT1 | 1443143_at | 10560 | 0.013 | -0.1139 | No | ||
| 80 | LRRTM3 | 1434759_at 1434760_at 1434761_at 1440112_at 1444477_at | 10581 | 0.013 | -0.1146 | No | ||
| 81 | UNC5B | 1435110_at 1453269_at | 10597 | 0.012 | -0.1151 | No | ||
| 82 | BARHL1 | 1418811_at 1418812_a_at | 10676 | 0.011 | -0.1186 | No | ||
| 83 | MYO1B | 1427450_x_at 1441210_at 1447364_x_at 1448989_a_at 1448990_a_at 1458007_at 1459679_s_at | 10957 | 0.005 | -0.1313 | No | ||
| 84 | ETV4 | 1423232_at 1443381_at | 11014 | 0.004 | -0.1339 | No | ||
| 85 | OTX2 | 1425926_a_at | 11024 | 0.003 | -0.1342 | No | ||
| 86 | CDKN2C | 1416868_at 1439164_at | 11227 | -0.000 | -0.1435 | No | ||
| 87 | FIGN | 1450268_at | 11275 | -0.002 | -0.1456 | No | ||
| 88 | OGG1 | 1430078_a_at 1448815_at | 11293 | -0.002 | -0.1464 | No | ||
| 89 | SLCO5A1 | 1439588_at 1440874_at | 11305 | -0.003 | -0.1469 | No | ||
| 90 | MYT1 | 1422773_at 1439365_at | 11463 | -0.006 | -0.1540 | No | ||
| 91 | MYOG | 1419391_at | 11468 | -0.006 | -0.1541 | No | ||
| 92 | AQP6 | 1420035_at 1441978_at 1447974_s_at | 11498 | -0.007 | -0.1553 | No | ||
| 93 | HOXC4 | 1422870_at | 11549 | -0.008 | -0.1575 | No | ||
| 94 | TGM3 | 1421355_at 1440150_at | 11699 | -0.011 | -0.1642 | No | ||
| 95 | SIX3 | 1426637_a_at 1426638_at | 11735 | -0.012 | -0.1657 | No | ||
| 96 | TITF1 | 1422346_at | 11840 | -0.014 | -0.1702 | No | ||
| 97 | FILIP1 | 1436650_at 1444075_at | 11988 | -0.017 | -0.1768 | No | ||
| 98 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 12009 | -0.017 | -0.1774 | No | ||
| 99 | SHOX2 | 1420559_a_at 1438042_at | 12033 | -0.018 | -0.1782 | No | ||
| 100 | PAK1IP1 | 1423766_at 1430875_a_at | 12180 | -0.021 | -0.1846 | No | ||
| 101 | DIXDC1 | 1425256_a_at 1435207_at 1443957_at 1444395_at | 12267 | -0.023 | -0.1883 | No | ||
| 102 | PNMA1 | 1429224_at | 12458 | -0.027 | -0.1966 | No | ||
| 103 | ATP2A3 | 1421129_a_at 1450124_a_at | 12654 | -0.031 | -0.2051 | No | ||
| 104 | HOXA10 | 1431475_a_at 1446408_at | 12703 | -0.032 | -0.2069 | No | ||
| 105 | SMARCA2 | 1430526_a_at 1439930_at 1452333_at | 12704 | -0.032 | -0.2064 | No | ||
| 106 | JPH3 | 1418161_at 1443724_at | 12754 | -0.034 | -0.2082 | No | ||
| 107 | CGN | 1430329_at 1435155_at | 12950 | -0.038 | -0.2166 | No | ||
| 108 | BNC2 | 1438861_at 1440593_at 1459132_at | 13142 | -0.042 | -0.2248 | No | ||
| 109 | PRDX5 | 1416381_a_at | 13254 | -0.045 | -0.2293 | No | ||
| 110 | DTNA | 1419223_a_at 1425292_at 1426066_a_at 1427588_a_at 1429768_at 1447280_at 1456069_at 1458908_at | 13298 | -0.045 | -0.2306 | No | ||
| 111 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 13829 | -0.058 | -0.2541 | No | ||
| 112 | NKX2-2 | 1421112_at | 13837 | -0.058 | -0.2536 | No | ||
| 113 | SLITRK1 | 1428089_at | 13875 | -0.059 | -0.2545 | No | ||
| 114 | DNAJB7 | 1450991_at | 14060 | -0.063 | -0.2620 | No | ||
| 115 | HESX1 | 1420604_at | 14151 | -0.066 | -0.2652 | No | ||
| 116 | NOL4 | 1430189_at 1441447_at 1456387_at | 14686 | -0.080 | -0.2886 | No | ||
| 117 | POLR2C | 1416341_at | 14947 | -0.087 | -0.2993 | No | ||
| 118 | BAD | 1416582_a_at 1416583_at | 15444 | -0.101 | -0.3206 | No | ||
| 119 | BTBD3 | 1425660_at 1433868_at 1442748_at | 15472 | -0.102 | -0.3204 | No | ||
| 120 | PRDM1 | 1420425_at 1444997_at | 15531 | -0.104 | -0.3216 | No | ||
| 121 | TBX6 | 1449868_at | 15787 | -0.112 | -0.3317 | No | ||
| 122 | LMO3 | 1455754_at | 15878 | -0.115 | -0.3342 | No | ||
| 123 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 15908 | -0.116 | -0.3339 | No | ||
| 124 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 16059 | -0.121 | -0.3391 | No | ||
| 125 | DAB1 | 1421100_a_at 1427307_a_at 1427308_at 1435577_at 1435578_s_at | 16137 | -0.123 | -0.3409 | No | ||
| 126 | NHLH2 | 1436888_at | 16164 | -0.124 | -0.3403 | No | ||
| 127 | SYNGR4 | 1418875_at | 16203 | -0.126 | -0.3402 | No | ||
| 128 | EYA1 | 1421727_at 1443117_at 1457424_at | 16261 | -0.128 | -0.3410 | No | ||
| 129 | GAL3ST4 | 1435606_at | 16423 | -0.133 | -0.3465 | No | ||
| 130 | MYST4 | 1423508_at 1446189_at 1447258_at 1447758_x_at | 16531 | -0.137 | -0.3495 | No | ||
| 131 | LUZP1 | 1416469_at 1448352_at | 16692 | -0.144 | -0.3548 | No | ||
| 132 | ACTR10 | 1417157_at | 16968 | -0.155 | -0.3652 | No | ||
| 133 | SLC7A9 | 1448783_at | 17129 | -0.162 | -0.3703 | No | ||
| 134 | DSCR1 | 1416600_a_at 1416601_a_at | 17441 | -0.178 | -0.3820 | No | ||
| 135 | IRS1 | 1423104_at | 17881 | -0.207 | -0.3992 | No | ||
| 136 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 17965 | -0.212 | -0.4000 | No | ||
| 137 | RTN4RL1 | 1436868_at 1455664_at | 18005 | -0.215 | -0.3988 | No | ||
| 138 | ZIC1 | 1423477_at 1439627_at | 18082 | -0.221 | -0.3991 | No | ||
| 139 | COL11A2 | 1423578_at | 18151 | -0.227 | -0.3990 | No | ||
| 140 | CCNG1 | 1420827_a_at 1450016_at 1450017_at | 18909 | -0.293 | -0.4296 | No | ||
| 141 | LGALS12 | 1417686_at 1417687_at | 19066 | -0.312 | -0.4323 | No | ||
| 142 | HOXA4 | 1420226_x_at 1427354_at | 19533 | -0.380 | -0.4483 | No | ||
| 143 | UNC13D | 1428992_at | 19659 | -0.400 | -0.4484 | Yes | ||
| 144 | ELMO1 | 1424523_at 1446610_at 1450208_a_at | 19716 | -0.409 | -0.4451 | Yes | ||
| 145 | EMID1 | 1442144_at 1449581_at | 19821 | -0.429 | -0.4438 | Yes | ||
| 146 | TLE3 | 1419654_at 1419655_at 1449554_at 1458512_at | 19828 | -0.431 | -0.4379 | Yes | ||
| 147 | FGF13 | 1418497_at 1418498_at | 19833 | -0.432 | -0.4319 | Yes | ||
| 148 | CHD2 | 1444246_at 1445843_at | 19913 | -0.449 | -0.4292 | Yes | ||
| 149 | CAP1 | 1417461_at 1417462_at | 20194 | -0.519 | -0.4346 | Yes | ||
| 150 | CDC42EP3 | 1422642_at 1450700_at | 20302 | -0.553 | -0.4317 | Yes | ||
| 151 | ITPR3 | 1417297_at | 20439 | -0.595 | -0.4294 | Yes | ||
| 152 | CHCHD7 | 1440158_x_at 1444318_at 1446844_at 1454640_at | 20620 | -0.661 | -0.4283 | Yes | ||
| 153 | SDPR | 1416778_at 1416779_at 1443832_s_at | 20719 | -0.710 | -0.4227 | Yes | ||
| 154 | NFIL3 | 1418932_at | 20737 | -0.724 | -0.4131 | Yes | ||
| 155 | NLGN2 | 1455143_at 1460013_at | 20745 | -0.728 | -0.4031 | Yes | ||
| 156 | PLAG1 | 1421745_at 1443943_at | 20954 | -0.857 | -0.4004 | Yes | ||
| 157 | CACNB3 | 1448656_at | 21008 | -0.896 | -0.3901 | Yes | ||
| 158 | SLC20A1 | 1438824_at 1446441_at 1448568_a_at | 21196 | -1.076 | -0.3834 | Yes | ||
| 159 | JARID1C | 1426497_at 1426498_at 1441449_at 1441450_s_at 1444157_a_at 1444158_at 1457930_at | 21295 | -1.184 | -0.3710 | Yes | ||
| 160 | UTX | 1427235_at 1427672_a_at 1440524_at 1441201_at 1445198_at 1446234_at 1446278_at | 21341 | -1.267 | -0.3550 | Yes | ||
| 161 | DLX1 | 1449470_at | 21371 | -1.298 | -0.3378 | Yes | ||
| 162 | CLDN4 | 1418283_at | 21416 | -1.371 | -0.3203 | Yes | ||
| 163 | GNAI1 | 1427510_at 1434440_at 1454959_s_at | 21426 | -1.386 | -0.3010 | Yes | ||
| 164 | TGFB3 | 1417455_at | 21503 | -1.541 | -0.2825 | Yes | ||
| 165 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 21658 | -1.921 | -0.2622 | Yes | ||
| 166 | STK38 | 1416252_at 1431303_at 1439938_at | 21695 | -2.083 | -0.2342 | Yes | ||
| 167 | LTBP3 | 1418049_at 1437833_at 1438312_s_at 1456189_x_at | 21698 | -2.090 | -0.2045 | Yes | ||
| 168 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 21702 | -2.099 | -0.1747 | Yes | ||
| 169 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 21799 | -2.661 | -0.1412 | Yes | ||
| 170 | IL5 | 1425546_a_at 1442049_at 1450550_at | 21895 | -4.125 | -0.0868 | Yes | ||
| 171 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 21930 | -6.219 | 0.0003 | Yes |