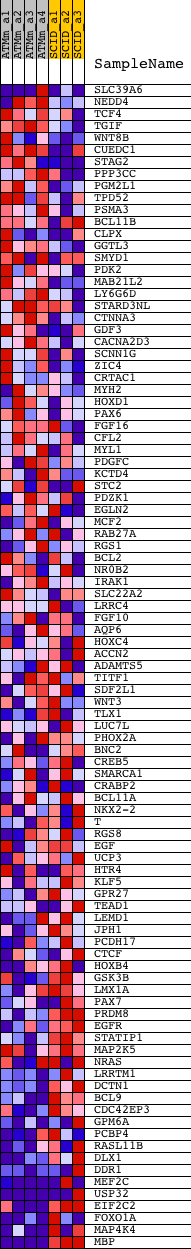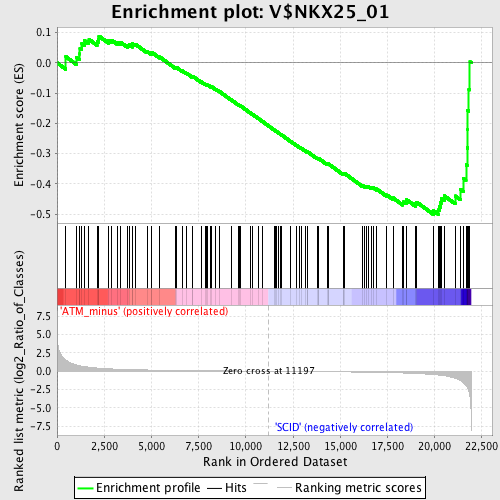
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | V$NKX25_01 |
| Enrichment Score (ES) | -0.50076294 |
| Normalized Enrichment Score (NES) | -1.4774057 |
| Nominal p-value | 0.011976048 |
| FDR q-value | 0.3530976 |
| FWER p-Value | 0.945 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SLC39A6 | 1424674_at 1424675_at 1441949_x_at 1447893_x_at | 464 | 1.539 | 0.0202 | No | ||
| 2 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 1036 | 0.857 | 0.0171 | No | ||
| 3 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 1203 | 0.741 | 0.0295 | No | ||
| 4 | TGIF | 1422286_a_at | 1210 | 0.737 | 0.0491 | No | ||
| 5 | WNT8B | 1421439_at | 1312 | 0.684 | 0.0629 | No | ||
| 6 | CUEDC1 | 1425920_at 1435358_at 1443455_at 1451447_at | 1461 | 0.628 | 0.0730 | No | ||
| 7 | STAG2 | 1421849_at 1450396_at | 1687 | 0.557 | 0.0777 | No | ||
| 8 | PPP3CC | 1420743_a_at 1430025_at | 2116 | 0.440 | 0.0700 | No | ||
| 9 | PGM2L1 | 1438774_s_at 1452841_at 1456478_at | 2175 | 0.429 | 0.0789 | No | ||
| 10 | TPD52 | 1419493_a_at 1419494_a_at 1436008_at 1458668_at | 2215 | 0.421 | 0.0884 | No | ||
| 11 | PSMA3 | 1448442_a_at | 2731 | 0.333 | 0.0738 | No | ||
| 12 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 2895 | 0.313 | 0.0748 | No | ||
| 13 | CLPX | 1423447_at 1439342_at | 3213 | 0.275 | 0.0676 | No | ||
| 14 | GGTL3 | 1451094_at | 3342 | 0.263 | 0.0689 | No | ||
| 15 | SMYD1 | 1421329_a_at 1427714_at 1441667_s_at 1443726_at 1443727_x_at 1450203_at | 3729 | 0.232 | 0.0574 | No | ||
| 16 | PDK2 | 1448825_at | 3807 | 0.225 | 0.0600 | No | ||
| 17 | MAB21L2 | 1418934_at | 3977 | 0.213 | 0.0580 | No | ||
| 18 | LY6G6D | 1450774_at | 3991 | 0.212 | 0.0631 | No | ||
| 19 | STARD3NL | 1430274_a_at | 4144 | 0.202 | 0.0616 | No | ||
| 20 | CTNNA3 | 1441560_at | 4773 | 0.168 | 0.0373 | No | ||
| 21 | GDF3 | 1449288_at 1460057_at | 4999 | 0.158 | 0.0313 | No | ||
| 22 | CACNA2D3 | 1419225_at | 5020 | 0.157 | 0.0346 | No | ||
| 23 | SCNN1G | 1419079_at | 5429 | 0.141 | 0.0197 | No | ||
| 24 | ZIC4 | 1421539_at 1456417_at | 6275 | 0.113 | -0.0159 | No | ||
| 25 | CRTAC1 | 1426606_at 1452936_at 1457474_at 1459858_x_at | 6299 | 0.112 | -0.0140 | No | ||
| 26 | MYH2 | 1425153_at | 6629 | 0.102 | -0.0263 | No | ||
| 27 | HOXD1 | 1420573_at | 6847 | 0.096 | -0.0336 | No | ||
| 28 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 7178 | 0.087 | -0.0464 | No | ||
| 29 | FGF16 | 1420806_at | 7182 | 0.087 | -0.0442 | No | ||
| 30 | CFL2 | 1418066_at 1418067_at 1431432_at | 7631 | 0.076 | -0.0627 | No | ||
| 31 | MYL1 | 1452651_a_at | 7832 | 0.071 | -0.0699 | No | ||
| 32 | PDGFC | 1419123_a_at 1449351_s_at | 7914 | 0.069 | -0.0717 | No | ||
| 33 | KCTD4 | 1420537_at 1441801_at | 7990 | 0.068 | -0.0734 | No | ||
| 34 | STC2 | 1419503_at 1445186_at 1449484_at | 8136 | 0.064 | -0.0783 | No | ||
| 35 | PDZK1 | 1431701_a_at | 8186 | 0.064 | -0.0788 | No | ||
| 36 | EGLN2 | 1416533_at | 8391 | 0.059 | -0.0866 | No | ||
| 37 | MCF2 | 1419021_at | 8604 | 0.053 | -0.0948 | No | ||
| 38 | RAB27A | 1425284_a_at 1425285_a_at 1429123_at 1454438_at | 9223 | 0.040 | -0.1221 | No | ||
| 39 | RGS1 | 1417601_at | 9603 | 0.032 | -0.1386 | No | ||
| 40 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 9673 | 0.030 | -0.1409 | No | ||
| 41 | NR0B2 | 1449854_at | 9692 | 0.030 | -0.1409 | No | ||
| 42 | IRAK1 | 1448668_a_at 1460649_at | 10222 | 0.020 | -0.1646 | No | ||
| 43 | SLC22A2 | 1419117_at | 10345 | 0.017 | -0.1697 | No | ||
| 44 | LRRC4 | 1416097_at 1435832_at | 10684 | 0.011 | -0.1849 | No | ||
| 45 | FGF10 | 1420690_at | 10893 | 0.006 | -0.1943 | No | ||
| 46 | AQP6 | 1420035_at 1441978_at 1447974_s_at | 11498 | -0.007 | -0.2218 | No | ||
| 47 | HOXC4 | 1422870_at | 11549 | -0.008 | -0.2239 | No | ||
| 48 | ACCN2 | 1455328_at | 11597 | -0.009 | -0.2258 | No | ||
| 49 | ADAMTS5 | 1422561_at 1450658_at 1456404_at | 11713 | -0.011 | -0.2307 | No | ||
| 50 | TITF1 | 1422346_at | 11840 | -0.014 | -0.2361 | No | ||
| 51 | SDF2L1 | 1418206_at | 11878 | -0.015 | -0.2374 | No | ||
| 52 | WNT3 | 1450763_x_at | 12376 | -0.025 | -0.2595 | No | ||
| 53 | TLX1 | 1450526_at | 12669 | -0.032 | -0.2720 | No | ||
| 54 | LUC7L | 1428255_at 1431181_a_at 1431664_at 1452708_a_at | 12832 | -0.035 | -0.2785 | No | ||
| 55 | PHOX2A | 1418620_at | 12922 | -0.037 | -0.2816 | No | ||
| 56 | BNC2 | 1438861_at 1440593_at 1459132_at | 13142 | -0.042 | -0.2905 | No | ||
| 57 | CREB5 | 1440089_at 1442576_at 1457222_at | 13244 | -0.044 | -0.2939 | No | ||
| 58 | SMARCA1 | 1460292_a_at | 13279 | -0.045 | -0.2942 | No | ||
| 59 | CRABP2 | 1451191_at | 13771 | -0.056 | -0.3152 | No | ||
| 60 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 13829 | -0.058 | -0.3163 | No | ||
| 61 | NKX2-2 | 1421112_at | 13837 | -0.058 | -0.3150 | No | ||
| 62 | T | 1419304_at | 14295 | -0.069 | -0.3341 | No | ||
| 63 | RGS8 | 1453060_at | 14358 | -0.071 | -0.3350 | No | ||
| 64 | EGF | 1418093_a_at | 14364 | -0.072 | -0.3333 | No | ||
| 65 | UCP3 | 1420657_at 1420658_at | 15147 | -0.092 | -0.3666 | No | ||
| 66 | HTR4 | 1427654_a_at 1439614_at 1443365_at | 15169 | -0.093 | -0.3651 | No | ||
| 67 | KLF5 | 1430255_at 1451021_a_at 1451739_at | 15226 | -0.095 | -0.3651 | No | ||
| 68 | GPR27 | 1422154_at | 16162 | -0.124 | -0.4045 | No | ||
| 69 | TEAD1 | 1423434_at 1447911_at 1456717_at | 16288 | -0.129 | -0.4068 | No | ||
| 70 | LEMD1 | 1445642_at | 16389 | -0.132 | -0.4078 | No | ||
| 71 | JPH1 | 1421520_at 1438428_at | 16496 | -0.136 | -0.4090 | No | ||
| 72 | PCDH17 | 1436920_at 1446821_at 1453070_at | 16660 | -0.143 | -0.4126 | No | ||
| 73 | CTCF | 1418330_at 1449042_at | 16736 | -0.146 | -0.4121 | No | ||
| 74 | HOXB4 | 1451761_at 1460379_at | 16918 | -0.152 | -0.4163 | No | ||
| 75 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 17459 | -0.179 | -0.4362 | No | ||
| 76 | LMX1A | 1421554_at | 17790 | -0.201 | -0.4459 | No | ||
| 77 | PAX7 | 1444596_at 1452510_at | 18308 | -0.237 | -0.4632 | No | ||
| 78 | PRDM8 | 1431852_at 1455925_at | 18365 | -0.242 | -0.4593 | No | ||
| 79 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 18510 | -0.256 | -0.4590 | No | ||
| 80 | STATIP1 | 1415774_at 1438179_s_at | 18514 | -0.257 | -0.4522 | No | ||
| 81 | MAP2K5 | 1417854_at 1430180_at 1453712_a_at 1455941_s_at | 18986 | -0.303 | -0.4656 | No | ||
| 82 | NRAS | 1422688_a_at 1454060_a_at | 19045 | -0.309 | -0.4599 | No | ||
| 83 | LRRTM1 | 1437746_at 1452624_at 1455883_a_at | 19938 | -0.455 | -0.4885 | Yes | ||
| 84 | DCTN1 | 1422521_at 1435414_s_at | 20182 | -0.517 | -0.4857 | Yes | ||
| 85 | BCL9 | 1445859_at 1446767_at 1451574_at 1455565_at | 20256 | -0.540 | -0.4745 | Yes | ||
| 86 | CDC42EP3 | 1422642_at 1450700_at | 20302 | -0.553 | -0.4617 | Yes | ||
| 87 | GPM6A | 1426442_at 1456741_s_at 1459303_at | 20340 | -0.562 | -0.4482 | Yes | ||
| 88 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20505 | -0.618 | -0.4391 | Yes | ||
| 89 | RASL11B | 1423854_a_at | 21084 | -0.964 | -0.4396 | Yes | ||
| 90 | DLX1 | 1449470_at | 21371 | -1.298 | -0.4177 | Yes | ||
| 91 | DDR1 | 1415797_at 1415798_at 1435820_x_at 1437619_x_at 1438367_x_at 1439382_x_at 1456226_x_at 1459990_at | 21518 | -1.579 | -0.3819 | Yes | ||
| 92 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 21658 | -1.921 | -0.3365 | Yes | ||
| 93 | USP32 | 1430403_at 1436159_at 1439857_at 1440399_at 1443689_at 1459857_at | 21718 | -2.179 | -0.2806 | Yes | ||
| 94 | EIF2C2 | 1426366_at 1438043_at | 21742 | -2.290 | -0.2199 | Yes | ||
| 95 | FOXO1A | 1416981_at 1416982_at 1416983_s_at 1459170_at | 21749 | -2.340 | -0.1572 | Yes | ||
| 96 | MAP4K4 | 1422615_at 1440609_at 1448050_s_at 1459912_at | 21811 | -2.707 | -0.0871 | Yes | ||
| 97 | MBP | 1419646_a_at 1425263_a_at 1425264_s_at 1433532_a_at 1436201_x_at 1451961_a_at 1454651_x_at 1456228_x_at | 21867 | -3.444 | 0.0032 | Yes |