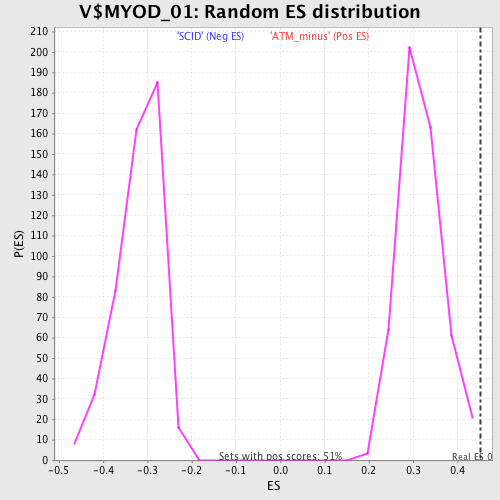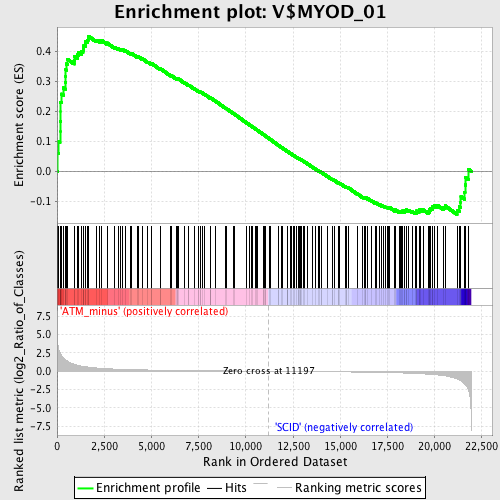
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | V$MYOD_01 |
| Enrichment Score (ES) | 0.45175835 |
| Normalized Enrichment Score (NES) | 1.4231342 |
| Nominal p-value | 0.0038910506 |
| FDR q-value | 0.51927936 |
| FWER p-Value | 0.996 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NDUFB8 | 1448198_a_at 1458404_at | 26 | 4.438 | 0.0610 | Yes | ||
| 2 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 93 | 2.929 | 0.0990 | Yes | ||
| 3 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 153 | 2.510 | 0.1315 | Yes | ||
| 4 | PTK2B | 1434653_at 1442437_at 1442927_at | 157 | 2.473 | 0.1660 | Yes | ||
| 5 | CACNB2 | 1441521_at 1444693_at 1446632_at 1452476_at 1456401_at | 179 | 2.373 | 0.1983 | Yes | ||
| 6 | CORO2A | 1445034_at 1452367_at | 181 | 2.371 | 0.2315 | Yes | ||
| 7 | FUT8 | 1441015_at 1442483_at 1443656_at 1456367_at 1460319_at | 241 | 2.103 | 0.2583 | Yes | ||
| 8 | EGR1 | 1417065_at | 337 | 1.769 | 0.2787 | Yes | ||
| 9 | CKM | 1417614_at | 432 | 1.596 | 0.2967 | Yes | ||
| 10 | DARS | 1423800_at | 454 | 1.556 | 0.3176 | Yes | ||
| 11 | POLR1D | 1447320_x_at 1448287_at 1451120_at 1459657_s_at | 462 | 1.542 | 0.3389 | Yes | ||
| 12 | EGLN1 | 1423785_at 1443389_at 1451110_at | 480 | 1.485 | 0.3589 | Yes | ||
| 13 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 555 | 1.348 | 0.3744 | Yes | ||
| 14 | ITSN1 | 1421192_a_at 1425899_a_at 1435884_at 1435885_s_at 1440637_at 1441034_at 1442637_at 1452338_s_at 1459621_at | 901 | 0.952 | 0.3719 | Yes | ||
| 15 | STIM1 | 1436945_x_at 1448320_at 1458460_at | 916 | 0.943 | 0.3845 | Yes | ||
| 16 | PAK2 | 1434250_at 1454887_at | 1070 | 0.831 | 0.3891 | Yes | ||
| 17 | CUGBP1 | 1423932_at 1425932_a_at 1426407_at 1426408_at 1427413_a_at | 1144 | 0.779 | 0.3966 | Yes | ||
| 18 | UBXD2 | 1426484_at 1426485_at 1426486_at | 1290 | 0.695 | 0.3997 | Yes | ||
| 19 | PSIP1 | 1417166_at 1442148_at 1460403_at | 1372 | 0.662 | 0.4053 | Yes | ||
| 20 | CENTB1 | 1434873_a_at 1434874_x_at 1437117_at 1455125_at | 1388 | 0.653 | 0.4137 | Yes | ||
| 21 | COL18A1 | 1418237_s_at 1426955_at | 1417 | 0.643 | 0.4215 | Yes | ||
| 22 | TNNT2 | 1418726_a_at 1424967_x_at 1440424_at | 1498 | 0.618 | 0.4264 | Yes | ||
| 23 | EXOSC3 | 1431502_a_at | 1514 | 0.612 | 0.4343 | Yes | ||
| 24 | SOST | 1421245_at 1436240_at 1450179_at | 1619 | 0.577 | 0.4376 | Yes | ||
| 25 | CLDN9 | 1439427_at 1450524_at | 1640 | 0.571 | 0.4447 | Yes | ||
| 26 | SPI1 | 1418747_at | 1660 | 0.564 | 0.4518 | Yes | ||
| 27 | PRKCQ | 1426044_a_at | 2083 | 0.447 | 0.4386 | No | ||
| 28 | TMEM17 | 1456332_at 1459997_s_at | 2235 | 0.416 | 0.4375 | No | ||
| 29 | ZRANB1 | 1415712_at 1427200_at 1436228_at | 2360 | 0.390 | 0.4373 | No | ||
| 30 | CALB1 | 1417504_at 1448738_at 1458836_at | 2650 | 0.345 | 0.4289 | No | ||
| 31 | RIMS2 | 1422809_at 1432102_at 1436470_at 1443215_at 1450761_s_at | 3060 | 0.292 | 0.4142 | No | ||
| 32 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 3229 | 0.274 | 0.4103 | No | ||
| 33 | KCNB1 | 1423179_at 1423180_at | 3370 | 0.261 | 0.4075 | No | ||
| 34 | MOSPD1 | 1424280_at | 3445 | 0.253 | 0.4076 | No | ||
| 35 | CRYZL1 | 1430546_at 1430547_s_at 1440045_at 1442574_at 1451473_a_at | 3617 | 0.239 | 0.4031 | No | ||
| 36 | MFAP2 | 1417359_at 1447833_x_at | 3878 | 0.220 | 0.3943 | No | ||
| 37 | DSCAM | 1419293_at 1431801_at 1441082_at 1449411_at | 3934 | 0.216 | 0.3948 | No | ||
| 38 | ITGA3 | 1421997_s_at 1455158_at 1460305_at | 4259 | 0.195 | 0.3826 | No | ||
| 39 | ARID5A | 1451340_at | 4300 | 0.192 | 0.3835 | No | ||
| 40 | CRLF1 | 1418476_at | 4507 | 0.181 | 0.3765 | No | ||
| 41 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 4810 | 0.167 | 0.3650 | No | ||
| 42 | PGF | 1418471_at | 4985 | 0.158 | 0.3592 | No | ||
| 43 | TUBA4 | 1417373_a_at 1417374_at 1417375_at | 5013 | 0.157 | 0.3602 | No | ||
| 44 | POFUT1 | 1443881_at 1450137_at 1456323_at | 5468 | 0.140 | 0.3413 | No | ||
| 45 | ELA3A | 1415883_a_at 1415884_at 1435611_x_at 1442170_at | 5483 | 0.139 | 0.3426 | No | ||
| 46 | GRID2 | 1421435_at 1421436_at 1435487_at 1437824_at 1443101_at 1446879_at 1458756_at 1459245_s_at | 6013 | 0.121 | 0.3200 | No | ||
| 47 | NTRK2 | 1420837_at 1420838_at 1435196_at 1435305_at 1437560_at 1446712_at | 6052 | 0.119 | 0.3199 | No | ||
| 48 | FES | 1427368_x_at 1452410_a_at | 6341 | 0.111 | 0.3082 | No | ||
| 49 | PAX9 | 1421246_at 1421247_at | 6375 | 0.110 | 0.3083 | No | ||
| 50 | KAZALD1 | 1436528_at | 6389 | 0.110 | 0.3092 | No | ||
| 51 | GNB3 | 1449159_at | 6430 | 0.108 | 0.3089 | No | ||
| 52 | SCG2 | 1450708_at | 6751 | 0.099 | 0.2956 | No | ||
| 53 | RXRG | 1418782_at | 6976 | 0.093 | 0.2866 | No | ||
| 54 | CHD6 | 1427384_at 1431047_at | 6979 | 0.092 | 0.2878 | No | ||
| 55 | KCNH5 | 1441742_at | 7273 | 0.085 | 0.2755 | No | ||
| 56 | KCNN3 | 1421632_at 1459308_at | 7487 | 0.079 | 0.2668 | No | ||
| 57 | GNG8 | 1457755_at | 7496 | 0.079 | 0.2675 | No | ||
| 58 | PSD | 1435780_at | 7575 | 0.077 | 0.2650 | No | ||
| 59 | DRD1IP | 1428813_a_at | 7601 | 0.077 | 0.2650 | No | ||
| 60 | HES6 | 1436050_x_at 1452021_a_at | 7684 | 0.075 | 0.2622 | No | ||
| 61 | SYT2 | 1420418_at 1440323_at 1449866_at | 7823 | 0.071 | 0.2569 | No | ||
| 62 | ABLIM3 | 1434013_at 1446069_at | 8123 | 0.065 | 0.2441 | No | ||
| 63 | CHRNG | 1427728_at 1449532_at 1452520_a_at | 8134 | 0.064 | 0.2445 | No | ||
| 64 | STC2 | 1419503_at 1445186_at 1449484_at | 8136 | 0.064 | 0.2454 | No | ||
| 65 | SLC6A7 | 1455469_at | 8385 | 0.059 | 0.2348 | No | ||
| 66 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 8912 | 0.046 | 0.2113 | No | ||
| 67 | GRIK5 | 1418784_at | 8959 | 0.045 | 0.2098 | No | ||
| 68 | CFL1 | 1448346_at 1455138_x_at | 9356 | 0.037 | 0.1921 | No | ||
| 69 | QTRTD1 | 1421320_a_at 1451199_at | 9404 | 0.036 | 0.1905 | No | ||
| 70 | FGF11 | 1421793_at 1439959_at | 10047 | 0.023 | 0.1613 | No | ||
| 71 | ARL4A | 1425411_at 1431429_a_at 1435092_at | 10180 | 0.020 | 0.1555 | No | ||
| 72 | CAV3 | 1418413_at | 10285 | 0.018 | 0.1510 | No | ||
| 73 | ALCAM | 1426300_at 1426301_at 1437466_at 1437467_at 1443086_at 1446720_at 1447040_at | 10347 | 0.017 | 0.1484 | No | ||
| 74 | SCN5A | 1422194_at 1458813_at | 10490 | 0.014 | 0.1421 | No | ||
| 75 | HMGN2 | 1433507_a_at | 10546 | 0.013 | 0.1397 | No | ||
| 76 | GABRQ | 1421536_at | 10638 | 0.012 | 0.1357 | No | ||
| 77 | GABRA3 | 1421263_at | 10920 | 0.006 | 0.1229 | No | ||
| 78 | FGF9 | 1420795_at 1438718_at 1445258_at | 10941 | 0.005 | 0.1220 | No | ||
| 79 | ERBB3 | 1434606_at 1452482_at | 10975 | 0.005 | 0.1206 | No | ||
| 80 | GCK | 1419146_a_at 1425303_at | 11021 | 0.003 | 0.1186 | No | ||
| 81 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 11028 | 0.003 | 0.1183 | No | ||
| 82 | PRELP | 1416321_s_at 1416322_at | 11269 | -0.002 | 0.1073 | No | ||
| 83 | LAMC2 | 1421279_at 1452505_at | 11322 | -0.003 | 0.1050 | No | ||
| 84 | ETV1 | 1422607_at 1445314_at 1450684_at | 11721 | -0.011 | 0.0869 | No | ||
| 85 | KCNE1L | 1416640_at | 11739 | -0.012 | 0.0862 | No | ||
| 86 | GDPD2 | 1429076_a_at | 11901 | -0.015 | 0.0791 | No | ||
| 87 | RAB26 | 1435500_at | 11956 | -0.016 | 0.0768 | No | ||
| 88 | WT1 | 1425995_s_at 1443221_at | 12195 | -0.021 | 0.0662 | No | ||
| 89 | SEMA3F | 1420508_at 1425840_a_at 1438947_x_at | 12196 | -0.021 | 0.0665 | No | ||
| 90 | FNDC5 | 1435115_at 1453135_at | 12201 | -0.021 | 0.0666 | No | ||
| 91 | VKORC1L1 | 1429092_at 1434916_at | 12374 | -0.025 | 0.0590 | No | ||
| 92 | TUBA1 | 1418884_x_at | 12437 | -0.026 | 0.0565 | No | ||
| 93 | EML1 | 1458317_at | 12504 | -0.028 | 0.0539 | No | ||
| 94 | NCDN | 1416852_a_at 1416853_at | 12589 | -0.030 | 0.0504 | No | ||
| 95 | HOXA10 | 1431475_a_at 1446408_at | 12703 | -0.032 | 0.0457 | No | ||
| 96 | GPR37 | 1450875_at | 12776 | -0.034 | 0.0429 | No | ||
| 97 | BACH1 | 1440831_at 1449311_at | 12794 | -0.034 | 0.0426 | No | ||
| 98 | LUC7L | 1428255_at 1431181_a_at 1431664_at 1452708_a_at | 12832 | -0.035 | 0.0414 | No | ||
| 99 | KCNK16 | 1429913_at | 12896 | -0.036 | 0.0390 | No | ||
| 100 | EFNA1 | 1416895_at 1448510_at | 12912 | -0.037 | 0.0388 | No | ||
| 101 | PIM2 | 1417216_at | 12962 | -0.038 | 0.0371 | No | ||
| 102 | DRP2 | 1440452_at 1441224_at 1441542_at | 13074 | -0.041 | 0.0326 | No | ||
| 103 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 13111 | -0.042 | 0.0315 | No | ||
| 104 | NHLH1 | 1419533_at | 13287 | -0.045 | 0.0241 | No | ||
| 105 | KIF9 | 1420395_a_at | 13533 | -0.051 | 0.0135 | No | ||
| 106 | TNFSF13 | 1445227_at | 13700 | -0.055 | 0.0067 | No | ||
| 107 | HRH3 | 1445285_at 1448807_at | 13868 | -0.059 | -0.0002 | No | ||
| 108 | RARA | 1450180_a_at | 13871 | -0.059 | 0.0006 | No | ||
| 109 | WNT6 | 1419708_at 1435980_x_at | 13997 | -0.062 | -0.0043 | No | ||
| 110 | ARRDC3 | 1454617_at | 14002 | -0.062 | -0.0036 | No | ||
| 111 | SRPK1 | 1454042_a_at 1455107_at | 14305 | -0.070 | -0.0165 | No | ||
| 112 | PPARGC1A | 1434099_at 1434100_x_at 1437751_at 1456395_at 1460336_at | 14560 | -0.077 | -0.0271 | No | ||
| 113 | MT3 | 1420575_at | 14567 | -0.077 | -0.0263 | No | ||
| 114 | PCDH1 | 1452294_at | 14700 | -0.081 | -0.0312 | No | ||
| 115 | SLC5A3 | 1422170_at | 14899 | -0.086 | -0.0391 | No | ||
| 116 | HTR2C | 1435513_at 1450477_at | 14910 | -0.086 | -0.0384 | No | ||
| 117 | CNNM2 | 1423475_at 1451007_at 1458132_at | 14960 | -0.088 | -0.0394 | No | ||
| 118 | PTK7 | 1452589_at | 15252 | -0.096 | -0.0514 | No | ||
| 119 | MUSK | 1450511_at | 15330 | -0.098 | -0.0536 | No | ||
| 120 | BTBD11 | 1428377_at 1459838_s_at 1459839_x_at | 15341 | -0.098 | -0.0527 | No | ||
| 121 | CACNA2D2 | 1426185_at | 15436 | -0.101 | -0.0556 | No | ||
| 122 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 15908 | -0.116 | -0.0756 | No | ||
| 123 | CYP26A1 | 1419430_at | 16183 | -0.125 | -0.0864 | No | ||
| 124 | HYAL2 | 1448679_at | 16297 | -0.129 | -0.0898 | No | ||
| 125 | UNC13B | 1417757_at 1443308_at | 16334 | -0.130 | -0.0897 | No | ||
| 126 | UBXD3 | 1426007_a_at 1451906_at 1456385_x_at 1457581_at | 16355 | -0.131 | -0.0887 | No | ||
| 127 | GCNT3 | 1424901_at 1430963_at | 16462 | -0.134 | -0.0917 | No | ||
| 128 | JMJD2C | 1424458_at 1430628_at 1447091_at | 16636 | -0.141 | -0.0977 | No | ||
| 129 | TSGA14 | 1419686_at 1434576_at 1443792_at 1445416_at | 16841 | -0.150 | -0.1050 | No | ||
| 130 | KTN1 | 1446702_at 1446749_at 1455434_a_at 1458502_at 1459429_at | 16912 | -0.152 | -0.1061 | No | ||
| 131 | ANKRD2 | 1419621_at | 17078 | -0.159 | -0.1114 | No | ||
| 132 | CALN1 | 1425846_a_at | 17204 | -0.165 | -0.1148 | No | ||
| 133 | SPOP | 1416524_at 1416525_at 1444578_at 1447827_x_at 1455510_at 1458886_at | 17294 | -0.170 | -0.1165 | No | ||
| 134 | RAC2 | 1417620_at 1440208_at | 17412 | -0.176 | -0.1194 | No | ||
| 135 | CSMD3 | 1438272_at 1438572_at 1454245_at | 17489 | -0.180 | -0.1204 | No | ||
| 136 | TMEM23 | 1426575_at 1426576_at 1436499_at 1442079_at 1445103_at 1457968_at | 17569 | -0.186 | -0.1214 | No | ||
| 137 | OMG | 1418212_at | 17589 | -0.188 | -0.1197 | No | ||
| 138 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 17892 | -0.208 | -0.1306 | No | ||
| 139 | CPNE1 | 1433439_at | 17920 | -0.209 | -0.1289 | No | ||
| 140 | COL11A2 | 1423578_at | 18151 | -0.227 | -0.1363 | No | ||
| 141 | ELF4 | 1421337_at 1445778_at | 18210 | -0.231 | -0.1358 | No | ||
| 142 | RBM3 | 1429169_at | 18224 | -0.232 | -0.1331 | No | ||
| 143 | CASKIN2 | 1450691_at | 18287 | -0.235 | -0.1327 | No | ||
| 144 | KCNN2 | 1445676_at 1448927_at | 18385 | -0.245 | -0.1337 | No | ||
| 145 | CAMK2A | 1437125_at 1441734_at 1442707_at 1443876_at 1452453_a_at 1457311_at | 18417 | -0.247 | -0.1317 | No | ||
| 146 | LNX2 | 1460249_at | 18488 | -0.253 | -0.1313 | No | ||
| 147 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 18516 | -0.257 | -0.1290 | No | ||
| 148 | PLXNC1 | 1423213_at 1423214_at 1443433_at 1443434_s_at 1450905_at 1450906_at | 18628 | -0.267 | -0.1303 | No | ||
| 149 | TAZ | 1416207_at 1453785_at | 18828 | -0.286 | -0.1355 | No | ||
| 150 | SOX12 | 1439754_at 1455562_at 1456882_at | 18989 | -0.304 | -0.1386 | No | ||
| 151 | CHCHD3 | 1431241_at 1437599_at 1451504_at 1459143_at | 19032 | -0.308 | -0.1362 | No | ||
| 152 | CBX8 | 1419198_at 1419199_at | 19051 | -0.310 | -0.1327 | No | ||
| 153 | MGAT1 | 1423609_a_at | 19169 | -0.327 | -0.1334 | No | ||
| 154 | USP54 | 1428731_at | 19176 | -0.328 | -0.1291 | No | ||
| 155 | SORCS2 | 1419358_at | 19269 | -0.342 | -0.1286 | No | ||
| 156 | ATOH7 | 1422929_s_at 1450796_at | 19394 | -0.359 | -0.1292 | No | ||
| 157 | EIF4ENIF1 | 1431075_a_at 1439169_at | 19653 | -0.398 | -0.1355 | No | ||
| 158 | ELMO1 | 1424523_at 1446610_at 1450208_a_at | 19716 | -0.409 | -0.1326 | No | ||
| 159 | TRIM37 | 1434554_at 1435604_at 1436393_a_at 1436394_at 1455012_s_at | 19720 | -0.409 | -0.1270 | No | ||
| 160 | EIF2C1 | 1434331_at | 19786 | -0.424 | -0.1241 | No | ||
| 161 | WBP2 | 1448121_at | 19872 | -0.440 | -0.1218 | No | ||
| 162 | CCNL2 | 1432195_s_at 1453740_a_at 1454149_a_at | 19890 | -0.444 | -0.1164 | No | ||
| 163 | SYT4 | 1415844_at 1415845_at | 19986 | -0.465 | -0.1142 | No | ||
| 164 | DDAH2 | 1416457_at | 20159 | -0.510 | -0.1150 | No | ||
| 165 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 20452 | -0.601 | -0.1200 | No | ||
| 166 | USP3 | 1425022_at 1425023_at 1435531_at 1441056_at 1446886_at | 20549 | -0.631 | -0.1155 | No | ||
| 167 | TNFRSF21 | 1422740_at 1450731_s_at | 21181 | -1.065 | -0.1296 | No | ||
| 168 | EPHB6 | 1418051_at | 21306 | -1.214 | -0.1183 | No | ||
| 169 | H2AFZ | 1441370_at | 21382 | -1.308 | -0.1034 | No | ||
| 170 | INVS | 1419308_at 1444709_at 1460491_at | 21389 | -1.318 | -0.0852 | No | ||
| 171 | ITGA6 | 1422444_at 1422445_at | 21588 | -1.745 | -0.0698 | No | ||
| 172 | DGKD | 1433564_at 1439873_at 1442254_at 1454655_at | 21605 | -1.783 | -0.0456 | No | ||
| 173 | CENTG3 | 1423910_at 1434032_at | 21622 | -1.824 | -0.0208 | No | ||
| 174 | PRICKLE1 | 1441663_at 1442172_at 1442400_at 1444759_at 1452249_at 1457637_at | 21775 | -2.507 | 0.0074 | No |