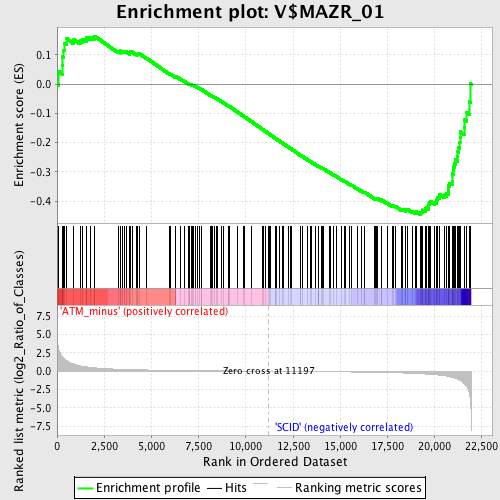
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | V$MAZR_01 |
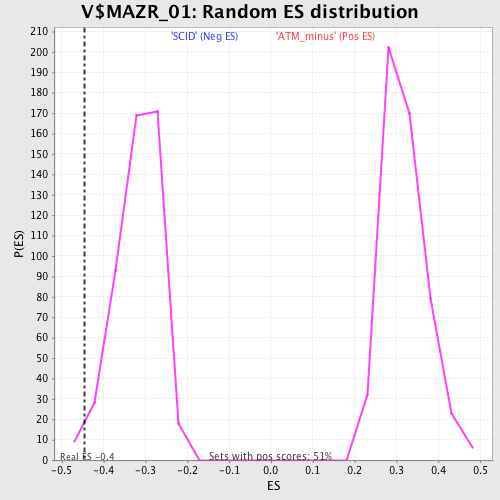
| Enrichment Score (ES) | -0.4457729 |
| Normalized Enrichment Score (NES) | -1.4048585 |
| Nominal p-value | 0.018442623 |
| FDR q-value | 0.40359682 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BAT2 | 1422799_at 1422800_at 1442802_x_at | 80 | 3.096 | 0.0430 | No | ||
| 2 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 263 | 2.018 | 0.0651 | No | ||
| 3 | UBE2R2 | 1415768_a_at 1443742_x_at 1444460_at | 283 | 1.939 | 0.0935 | No | ||
| 4 | SON | 1420951_a_at 1420952_at 1435862_at 1437924_at 1438871_at 1439074_a_at 1455634_at 1456630_x_at 1457596_at | 359 | 1.722 | 0.1160 | No | ||
| 5 | FGF17 | 1421523_at 1456239_at | 379 | 1.682 | 0.1405 | No | ||
| 6 | HNRPDL | 1420093_s_at 1420094_at 1424251_a_at 1424252_at 1428224_at 1428225_s_at 1449039_a_at 1456698_s_at | 516 | 1.416 | 0.1556 | No | ||
| 7 | EZH2 | 1416544_at 1444263_at | 891 | 0.961 | 0.1529 | No | ||
| 8 | ATF1 | 1417296_at | 1218 | 0.735 | 0.1490 | No | ||
| 9 | PARD6G | 1420851_at 1450025_at | 1351 | 0.673 | 0.1531 | No | ||
| 10 | TCEB1 | 1427914_a_at 1427915_s_at 1436226_at 1452593_a_at | 1536 | 0.605 | 0.1538 | No | ||
| 11 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 1574 | 0.592 | 0.1610 | No | ||
| 12 | CORO1C | 1417752_at 1419911_at 1437721_at 1442972_at 1449660_s_at 1459169_at | 1778 | 0.532 | 0.1597 | No | ||
| 13 | HMGA1 | 1416184_s_at | 1956 | 0.481 | 0.1588 | No | ||
| 14 | PIM1 | 1423006_at 1435458_at | 1987 | 0.474 | 0.1646 | No | ||
| 15 | CTNNBIP1 | 1417567_at 1431694_a_at | 3233 | 0.273 | 0.1115 | No | ||
| 16 | TLK2 | 1416944_a_at 1431827_a_at 1454018_at 1457357_at | 3339 | 0.264 | 0.1107 | No | ||
| 17 | CLCN6 | 1422314_at 1455440_at | 3355 | 0.262 | 0.1139 | No | ||
| 18 | CDC27 | 1426076_at 1433848_at 1433849_at 1454739_at | 3470 | 0.250 | 0.1125 | No | ||
| 19 | NEUROD2 | 1444362_at | 3570 | 0.242 | 0.1116 | No | ||
| 20 | ENTPD1 | 1423326_at 1450939_at 1453586_at | 3656 | 0.236 | 0.1112 | No | ||
| 21 | HSD11B1 | 1449038_at | 3834 | 0.223 | 0.1065 | No | ||
| 22 | MLL5 | 1427236_a_at 1432601_at 1434704_at 1439107_a_at 1439108_at | 3841 | 0.223 | 0.1096 | No | ||
| 23 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 3880 | 0.220 | 0.1111 | No | ||
| 24 | DUSP8 | 1418714_at | 3969 | 0.214 | 0.1103 | No | ||
| 25 | RTN2 | 1419056_at | 4200 | 0.198 | 0.1028 | No | ||
| 26 | AHR | 1422631_at 1450695_at | 4263 | 0.194 | 0.1028 | No | ||
| 27 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 4282 | 0.193 | 0.1049 | No | ||
| 28 | RBL1 | 1424156_at 1425166_at 1432604_at 1454293_at | 4340 | 0.190 | 0.1052 | No | ||
| 29 | HOXB6 | 1451660_a_at | 4752 | 0.170 | 0.0889 | No | ||
| 30 | FOXA1 | 1418496_at | 5943 | 0.123 | 0.0361 | No | ||
| 31 | GART | 1416283_at 1424435_a_at 1424436_at 1441911_x_at | 6011 | 0.121 | 0.0348 | No | ||
| 32 | SALL1 | 1450489_at | 6246 | 0.113 | 0.0258 | No | ||
| 33 | FXYD1 | 1421374_a_at | 6268 | 0.113 | 0.0265 | No | ||
| 34 | ZIC4 | 1421539_at 1456417_at | 6275 | 0.113 | 0.0279 | No | ||
| 35 | JPH4 | 1439031_at | 6529 | 0.105 | 0.0179 | No | ||
| 36 | TCF1 | 1421234_at | 6748 | 0.099 | 0.0094 | No | ||
| 37 | HOXB9 | 1452317_at | 6941 | 0.093 | 0.0020 | No | ||
| 38 | LIN28 | 1437752_at | 7006 | 0.092 | 0.0004 | No | ||
| 39 | SF1 | 1422321_a_at 1423750_a_at 1423751_at 1459765_s_at 1459766_x_at | 7034 | 0.091 | 0.0006 | No | ||
| 40 | HMX1 | 1422152_at | 7136 | 0.088 | -0.0028 | No | ||
| 41 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 7178 | 0.087 | -0.0033 | No | ||
| 42 | LRRTM4 | 1437214_at 1455937_at | 7231 | 0.086 | -0.0044 | No | ||
| 43 | LRRN6A | 1428118_at 1437386_at | 7327 | 0.084 | -0.0075 | No | ||
| 44 | POU3F2 | 1450831_at 1453478_at 1457734_at | 7428 | 0.081 | -0.0109 | No | ||
| 45 | LRFN5 | 1441502_at 1454687_at | 7544 | 0.078 | -0.0150 | No | ||
| 46 | SLC39A2 | 1443163_at | 7621 | 0.076 | -0.0173 | No | ||
| 47 | WNT1 | 1425377_at | 7654 | 0.075 | -0.0177 | No | ||
| 48 | STC2 | 1419503_at 1445186_at 1449484_at | 8136 | 0.064 | -0.0388 | No | ||
| 49 | MTHFR | 1422132_at 1434087_at 1450498_at | 8171 | 0.064 | -0.0394 | No | ||
| 50 | GRK5 | 1446361_at 1446998_at 1449514_at | 8221 | 0.063 | -0.0407 | No | ||
| 51 | THPO | 1449569_at | 8338 | 0.060 | -0.0451 | No | ||
| 52 | MAGED2 | 1426306_a_at 1459873_x_at | 8420 | 0.058 | -0.0480 | No | ||
| 53 | EIF5A | 1437859_x_at 1451470_s_at | 8504 | 0.055 | -0.0509 | No | ||
| 54 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 8690 | 0.051 | -0.0587 | No | ||
| 55 | MBD3 | 1417728_at | 8788 | 0.049 | -0.0624 | No | ||
| 56 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 9075 | 0.043 | -0.0749 | No | ||
| 57 | SYT3 | 1417708_at | 9090 | 0.042 | -0.0749 | No | ||
| 58 | FXYD3 | 1418374_at | 9123 | 0.042 | -0.0757 | No | ||
| 59 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 9575 | 0.032 | -0.0959 | No | ||
| 60 | RAB11B | 1423448_at | 9861 | 0.026 | -0.1086 | No | ||
| 61 | CAMK2G | 1423941_at 1423942_a_at | 9900 | 0.026 | -0.1100 | No | ||
| 62 | GGN | 1445278_at | 10291 | 0.018 | -0.1276 | No | ||
| 63 | SRF | 1418255_s_at 1418256_at | 10853 | 0.007 | -0.1533 | No | ||
| 64 | SEPT3 | 1417414_at 1439833_at 1444778_at | 10925 | 0.006 | -0.1565 | No | ||
| 65 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 11028 | 0.003 | -0.1611 | No | ||
| 66 | SLC39A5 | 1429523_a_at | 11207 | -0.000 | -0.1693 | No | ||
| 67 | NXPH4 | 1429753_at | 11264 | -0.002 | -0.1718 | No | ||
| 68 | SULT2B1 | 1417335_at | 11314 | -0.003 | -0.1740 | No | ||
| 69 | HOXC4 | 1422870_at | 11549 | -0.008 | -0.1847 | No | ||
| 70 | CACNB1 | 1425777_at 1426108_s_at 1440940_at 1451834_at | 11551 | -0.008 | -0.1846 | No | ||
| 71 | DLL3 | 1449236_at | 11627 | -0.009 | -0.1879 | No | ||
| 72 | APBA1 | 1437940_at 1455369_at 1459605_at | 11783 | -0.013 | -0.1948 | No | ||
| 73 | NOTCH2 | 1451889_at 1455556_at | 11948 | -0.016 | -0.2021 | No | ||
| 74 | FBXL19 | 1435922_at | 11966 | -0.016 | -0.2026 | No | ||
| 75 | PLA2G2E | 1420674_at | 12266 | -0.023 | -0.2160 | No | ||
| 76 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 12371 | -0.025 | -0.2204 | No | ||
| 77 | MYBPH | 1419487_at | 12400 | -0.026 | -0.2213 | No | ||
| 78 | HNRPUL1 | 1451984_at | 12910 | -0.037 | -0.2442 | No | ||
| 79 | EFNA1 | 1416895_at 1448510_at | 12912 | -0.037 | -0.2436 | No | ||
| 80 | FBS1 | 1425276_at 1451645_at | 12998 | -0.039 | -0.2470 | No | ||
| 81 | BAI2 | 1435558_at | 13286 | -0.045 | -0.2595 | No | ||
| 82 | THRAP1 | 1436904_at 1438725_at 1444068_at 1446840_at 1453160_at 1453161_at | 13428 | -0.048 | -0.2652 | No | ||
| 83 | AEBP2 | 1434080_at 1435070_at 1437743_at | 13491 | -0.050 | -0.2673 | No | ||
| 84 | NCOA2 | 1435233_at 1435234_at 1446827_at 1450458_at | 13680 | -0.054 | -0.2751 | No | ||
| 85 | UBE4B | 1432499_a_at 1444465_at 1448410_at | 13841 | -0.058 | -0.2816 | No | ||
| 86 | HPCA | 1425833_a_at 1450930_at | 13847 | -0.058 | -0.2809 | No | ||
| 87 | NETO1 | 1425132_at 1456283_at | 13853 | -0.058 | -0.2803 | No | ||
| 88 | PACSIN3 | 1416995_at 1431534_at 1437834_s_at 1456191_x_at | 13992 | -0.062 | -0.2857 | No | ||
| 89 | BAZ2A | 1427209_at 1427210_at 1438192_s_at | 14054 | -0.063 | -0.2875 | No | ||
| 90 | NFYC | 1448963_at | 14082 | -0.064 | -0.2878 | No | ||
| 91 | TRIP10 | 1418092_s_at | 14402 | -0.073 | -0.3014 | No | ||
| 92 | GATA6 | 1425463_at 1425464_at 1443081_at | 14454 | -0.074 | -0.3026 | No | ||
| 93 | GRIN2D | 1421393_at | 14655 | -0.079 | -0.3106 | No | ||
| 94 | ZNF462 | 1439106_at 1456789_at | 14804 | -0.084 | -0.3161 | No | ||
| 95 | DLL4 | 1421826_at 1421827_at | 15067 | -0.090 | -0.3268 | No | ||
| 96 | FUS | 1451285_at 1451286_s_at 1455831_at 1458801_at | 15206 | -0.095 | -0.3317 | No | ||
| 97 | HR | 1422110_at 1435950_at | 15248 | -0.096 | -0.3321 | No | ||
| 98 | FGF14 | 1435747_at 1447221_x_at 1447480_at 1449958_a_at 1458830_at 1459073_x_at | 15505 | -0.103 | -0.3424 | No | ||
| 99 | SLITRK5 | 1429718_at | 15602 | -0.106 | -0.3452 | No | ||
| 100 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 15913 | -0.116 | -0.3576 | No | ||
| 101 | SLC22A17 | 1448209_a_at | 16114 | -0.123 | -0.3650 | No | ||
| 102 | EYA1 | 1421727_at 1443117_at 1457424_at | 16261 | -0.128 | -0.3698 | No | ||
| 103 | ZBTB26 | 1434629_at 1447721_at | 16289 | -0.129 | -0.3691 | No | ||
| 104 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 16818 | -0.149 | -0.3911 | No | ||
| 105 | FOXJ2 | 1420373_at 1420374_at | 16843 | -0.150 | -0.3899 | No | ||
| 106 | PTPRF | 1420841_at 1420842_at 1420843_at | 16931 | -0.153 | -0.3916 | No | ||
| 107 | SLC2A4 | 1415958_at 1415959_at | 16980 | -0.155 | -0.3915 | No | ||
| 108 | NTN2L | 1422056_at | 17163 | -0.164 | -0.3974 | No | ||
| 109 | RAB1B | 1424064_at | 17178 | -0.164 | -0.3955 | No | ||
| 110 | NPTX1 | 1422130_at 1434877_at | 17476 | -0.180 | -0.4065 | No | ||
| 111 | RFX1 | 1422109_at 1436059_at | 17746 | -0.197 | -0.4158 | No | ||
| 112 | DMPK | 1422989_a_at 1434944_at | 17812 | -0.202 | -0.4158 | No | ||
| 113 | MKNK2 | 1418300_a_at 1449029_at | 17912 | -0.209 | -0.4172 | No | ||
| 114 | RAB5C | 1424684_at | 18213 | -0.231 | -0.4275 | No | ||
| 115 | IL1RAPL1 | 1445821_at 1458964_at | 18289 | -0.235 | -0.4274 | No | ||
| 116 | MOV10 | 1416380_at 1447376_at | 18453 | -0.250 | -0.4311 | No | ||
| 117 | TRPS1 | 1434286_at 1438214_at 1440026_at 1441333_at 1443161_at 1449530_at 1456983_at 1457445_at | 18472 | -0.252 | -0.4281 | No | ||
| 118 | RNF111 | 1423304_a_at | 18551 | -0.260 | -0.4278 | No | ||
| 119 | DNMT3A | 1423063_at 1423064_at 1423065_at 1423066_at 1442309_at 1460324_at | 18801 | -0.284 | -0.4349 | No | ||
| 120 | JUNB | 1415899_at | 18963 | -0.300 | -0.4378 | No | ||
| 121 | LUC7L2 | 1423251_at 1427087_at 1436165_at 1436766_at 1436767_at 1437761_at 1458423_at | 19031 | -0.308 | -0.4362 | No | ||
| 122 | PHF12 | 1434922_at 1453579_at | 19240 | -0.337 | -0.4407 | Yes | ||
| 123 | ABR | 1433477_at | 19283 | -0.344 | -0.4374 | Yes | ||
| 124 | VASP | 1451097_at | 19337 | -0.351 | -0.4346 | Yes | ||
| 125 | KCNK6 | 1435342_at | 19362 | -0.354 | -0.4303 | Yes | ||
| 126 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 19501 | -0.374 | -0.4310 | Yes | ||
| 127 | EPHB2 | 1425015_at 1425016_at 1442471_at 1445417_at 1454022_at | 19516 | -0.377 | -0.4260 | Yes | ||
| 128 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 19585 | -0.388 | -0.4233 | Yes | ||
| 129 | SEMA4C | 1433920_at | 19675 | -0.403 | -0.4213 | Yes | ||
| 130 | GNB2 | 1450623_at | 19688 | -0.404 | -0.4157 | Yes | ||
| 131 | SP1 | 1418180_at 1448994_at 1454852_at | 19692 | -0.405 | -0.4098 | Yes | ||
| 132 | CLOCK | 1418659_at 1435775_at 1439314_at | 19713 | -0.408 | -0.4045 | Yes | ||
| 133 | RGL2 | 1450688_at | 19760 | -0.419 | -0.4003 | Yes | ||
| 134 | FKBP2 | 1450694_at | 20007 | -0.471 | -0.4045 | Yes | ||
| 135 | LGALS1 | 1419573_a_at 1455439_a_at | 20105 | -0.497 | -0.4015 | Yes | ||
| 136 | MBD6 | 1418375_at | 20116 | -0.500 | -0.3944 | Yes | ||
| 137 | DDAH2 | 1416457_at | 20159 | -0.510 | -0.3886 | Yes | ||
| 138 | KCND3 | 1426070_a_at 1435720_at 1442555_at 1447513_at | 20235 | -0.533 | -0.3840 | Yes | ||
| 139 | TRERF1 | 1444654_at | 20236 | -0.533 | -0.3760 | Yes | ||
| 140 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 20505 | -0.618 | -0.3790 | Yes | ||
| 141 | CBFB | 1442617_at 1460716_a_at | 20606 | -0.654 | -0.3737 | Yes | ||
| 142 | GRIK1 | 1427676_a_at 1439987_at | 20717 | -0.709 | -0.3681 | Yes | ||
| 143 | RKHD3 | 1437152_at | 20740 | -0.725 | -0.3582 | Yes | ||
| 144 | NLGN2 | 1455143_at 1460013_at | 20745 | -0.728 | -0.3474 | Yes | ||
| 145 | SLC4A2 | 1416637_at | 20790 | -0.748 | -0.3381 | Yes | ||
| 146 | MTF1 | 1418333_at 1428979_at 1429170_a_at 1440414_at 1452990_at | 20919 | -0.831 | -0.3315 | Yes | ||
| 147 | H1F0 | 1423702_at 1450522_a_at | 20925 | -0.835 | -0.3191 | Yes | ||
| 148 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20926 | -0.835 | -0.3065 | Yes | ||
| 149 | TRIM41 | 1424275_s_at 1440268_at | 20988 | -0.880 | -0.2960 | Yes | ||
| 150 | POGZ | 1434650_at 1454827_at 1455046_a_at 1457748_at | 20998 | -0.889 | -0.2830 | Yes | ||
| 151 | TNPO2 | 1425592_at 1425593_at 1437283_at | 21057 | -0.935 | -0.2716 | Yes | ||
| 152 | SSBP2 | 1429951_at 1438423_at 1449815_a_at 1451542_at | 21095 | -0.976 | -0.2586 | Yes | ||
| 153 | ENSA | 1423330_at 1428620_at | 21215 | -1.088 | -0.2476 | Yes | ||
| 154 | UBE1 | 1442767_s_at 1448116_at | 21227 | -1.105 | -0.2315 | Yes | ||
| 155 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 21245 | -1.128 | -0.2152 | Yes | ||
| 156 | KCNH2 | 1449544_a_at | 21329 | -1.249 | -0.2002 | Yes | ||
| 157 | UTX | 1427235_at 1427672_a_at 1440524_at 1441201_at 1445198_at 1446234_at 1446278_at | 21341 | -1.267 | -0.1816 | Yes | ||
| 158 | HNRPR | 1427129_a_at 1438807_at 1452030_a_at | 21352 | -1.282 | -0.1627 | Yes | ||
| 159 | CLIC1 | 1416656_at 1458665_at | 21564 | -1.653 | -0.1475 | Yes | ||
| 160 | ITGA6 | 1422444_at 1422445_at | 21588 | -1.745 | -0.1222 | Yes | ||
| 161 | UST | 1437293_x_at 1439218_at 1444275_at 1458897_at 1459970_at | 21683 | -2.014 | -0.0962 | Yes | ||
| 162 | GTPBP2 | 1416690_at 1416691_at 1442305_at 1448437_a_at | 21829 | -2.852 | -0.0598 | Yes | ||
| 163 | RTN3 | 1418101_a_at 1425539_a_at 1442365_at 1443220_at | 21901 | -4.288 | 0.0016 | Yes |