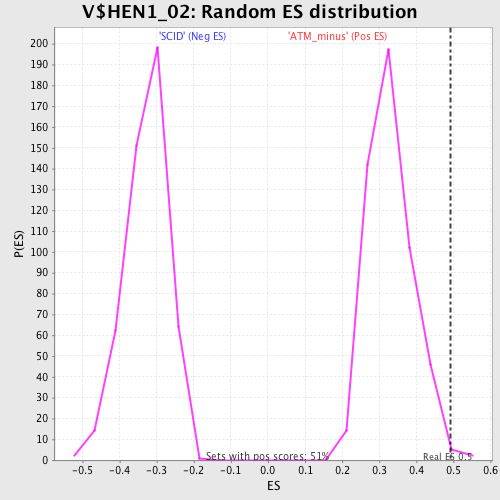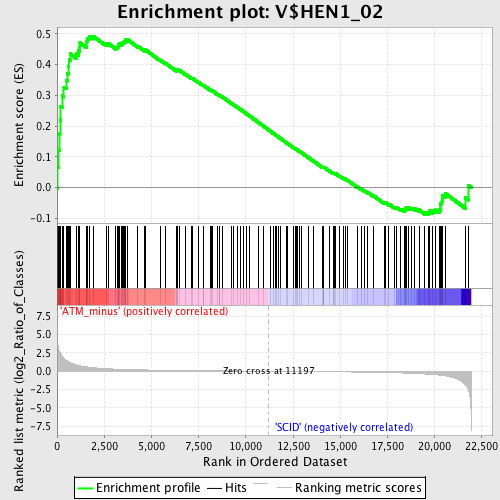
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | V$HEN1_02 |
| Enrichment Score (ES) | 0.49285302 |
| Normalized Enrichment Score (NES) | 1.4949567 |
| Nominal p-value | 0.005905512 |
| FDR q-value | 0.42796955 |
| FWER p-Value | 0.921 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CBX4 | 1419583_at 1440479_at | 43 | 3.624 | 0.0671 | Yes | ||
| 2 | EIF4G2 | 1415863_at 1452758_s_at | 85 | 3.012 | 0.1226 | Yes | ||
| 3 | NR4A3 | 1421079_at 1421080_at | 108 | 2.785 | 0.1746 | Yes | ||
| 4 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 153 | 2.510 | 0.2204 | Yes | ||
| 5 | CACNB2 | 1441521_at 1444693_at 1446632_at 1452476_at 1456401_at | 179 | 2.373 | 0.2645 | Yes | ||
| 6 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 263 | 2.018 | 0.2991 | Yes | ||
| 7 | DOCK4 | 1431114_at 1436405_at 1441462_at 1441469_at 1446283_at 1457146_at 1459279_at | 362 | 1.718 | 0.3273 | Yes | ||
| 8 | NR4A2 | 1447863_s_at 1450749_a_at 1450750_a_at 1455034_at | 491 | 1.465 | 0.3494 | Yes | ||
| 9 | ENO3 | 1417951_at 1449631_at | 548 | 1.355 | 0.3726 | Yes | ||
| 10 | PPP1R16B | 1425414_at 1455080_at | 617 | 1.245 | 0.3932 | Yes | ||
| 11 | PHLDB1 | 1424467_at 1424468_s_at | 629 | 1.231 | 0.4162 | Yes | ||
| 12 | ZFP91 | 1426326_at 1429615_at 1429616_at 1459444_at | 691 | 1.154 | 0.4353 | Yes | ||
| 13 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 1036 | 0.857 | 0.4359 | Yes | ||
| 14 | CUGBP1 | 1423932_at 1425932_a_at 1426407_at 1426408_at 1427413_a_at | 1144 | 0.779 | 0.4458 | Yes | ||
| 15 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 1203 | 0.741 | 0.4573 | Yes | ||
| 16 | TGIF | 1422286_a_at | 1210 | 0.737 | 0.4710 | Yes | ||
| 17 | FXC1 | 1417916_a_at 1436416_x_at 1448887_x_at 1453461_at | 1572 | 0.592 | 0.4658 | Yes | ||
| 18 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 1574 | 0.592 | 0.4770 | Yes | ||
| 19 | SOST | 1421245_at 1436240_at 1450179_at | 1619 | 0.577 | 0.4860 | Yes | ||
| 20 | RAB1A | 1416082_at 1448210_at 1457537_at | 1711 | 0.550 | 0.4923 | Yes | ||
| 21 | MYC | 1424942_a_at | 1905 | 0.494 | 0.4929 | Yes | ||
| 22 | BCL9L | 1419180_at 1438806_at 1443700_at 1444855_at | 2594 | 0.353 | 0.4680 | No | ||
| 23 | ASB2 | 1428444_at 1436259_at | 2703 | 0.337 | 0.4695 | No | ||
| 24 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 3117 | 0.285 | 0.4560 | No | ||
| 25 | ARFIP2 | 1424240_at 1435498_at | 3205 | 0.276 | 0.4573 | No | ||
| 26 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 3229 | 0.274 | 0.4614 | No | ||
| 27 | CRB1 | 1421451_at 1441330_at | 3263 | 0.270 | 0.4650 | No | ||
| 28 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 3297 | 0.267 | 0.4686 | No | ||
| 29 | CCNE2 | 1422535_at | 3419 | 0.255 | 0.4679 | No | ||
| 30 | MOSPD1 | 1424280_at | 3445 | 0.253 | 0.4716 | No | ||
| 31 | DPF3 | 1441659_at 1442686_at 1450162_at | 3524 | 0.246 | 0.4727 | No | ||
| 32 | NEUROD2 | 1444362_at | 3570 | 0.242 | 0.4752 | No | ||
| 33 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 3626 | 0.238 | 0.4772 | No | ||
| 34 | LOXL1 | 1436063_at 1451978_at | 3637 | 0.237 | 0.4813 | No | ||
| 35 | SLC29A4 | 1424900_at | 3740 | 0.230 | 0.4810 | No | ||
| 36 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 4282 | 0.193 | 0.4599 | No | ||
| 37 | NRG1 | 1456524_at | 4630 | 0.175 | 0.4473 | No | ||
| 38 | CHX10 | 1419628_at | 4671 | 0.173 | 0.4488 | No | ||
| 39 | POFUT1 | 1443881_at 1450137_at 1456323_at | 5468 | 0.140 | 0.4149 | No | ||
| 40 | AMPD2 | 1426757_at 1438941_x_at | 5734 | 0.130 | 0.4053 | No | ||
| 41 | RPIA | 1418337_at | 6337 | 0.111 | 0.3798 | No | ||
| 42 | FES | 1427368_x_at 1452410_a_at | 6341 | 0.111 | 0.3817 | No | ||
| 43 | ANK2 | 1434264_at 1434265_s_at 1440042_at 1440043_at 1443548_at 1444023_at 1445346_at 1446108_at 1459317_at | 6354 | 0.110 | 0.3833 | No | ||
| 44 | MYO18B | 1431813_at | 6395 | 0.109 | 0.3836 | No | ||
| 45 | DAAM1 | 1431035_at 1455244_at 1458662_at | 6477 | 0.106 | 0.3819 | No | ||
| 46 | TBX3 | 1437479_x_at 1439567_at 1444594_at 1445757_at 1448029_at | 6809 | 0.097 | 0.3685 | No | ||
| 47 | NLGN3 | 1456384_at | 7121 | 0.089 | 0.3560 | No | ||
| 48 | BCL6B | 1418421_at | 7165 | 0.088 | 0.3557 | No | ||
| 49 | GNG8 | 1457755_at | 7496 | 0.079 | 0.3420 | No | ||
| 50 | ALX3 | 1420555_at 1459790_x_at | 7730 | 0.073 | 0.3328 | No | ||
| 51 | HOXB5 | 1418415_at | 7755 | 0.073 | 0.3330 | No | ||
| 52 | KLHL1 | 1436465_at 1449241_at | 8107 | 0.065 | 0.3182 | No | ||
| 53 | SOX14 | 1427609_at | 8174 | 0.064 | 0.3164 | No | ||
| 54 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 8247 | 0.062 | 0.3143 | No | ||
| 55 | ASB16 | 1452023_at | 8508 | 0.055 | 0.3034 | No | ||
| 56 | MEGF10 | 1429841_at | 8580 | 0.054 | 0.3011 | No | ||
| 57 | PTGDS | 1423859_a_at 1423860_at | 8609 | 0.053 | 0.3009 | No | ||
| 58 | CREB3L1 | 1419295_at | 8753 | 0.050 | 0.2953 | No | ||
| 59 | RAB27A | 1425284_a_at 1425285_a_at 1429123_at 1454438_at | 9223 | 0.040 | 0.2745 | No | ||
| 60 | STMN2 | 1423280_at 1423281_at | 9350 | 0.037 | 0.2694 | No | ||
| 61 | CHD4 | 1436343_at 1438476_a_at 1451295_a_at | 9575 | 0.032 | 0.2598 | No | ||
| 62 | MFAP4 | 1424010_at | 9693 | 0.030 | 0.2550 | No | ||
| 63 | UNC119 | 1418123_at | 9866 | 0.026 | 0.2476 | No | ||
| 64 | LRRN3 | 1434539_at | 10026 | 0.023 | 0.2407 | No | ||
| 65 | ARL4A | 1425411_at 1431429_a_at 1435092_at | 10180 | 0.020 | 0.2341 | No | ||
| 66 | TRIM46 | 1460568_at | 10641 | 0.012 | 0.2132 | No | ||
| 67 | SEPT3 | 1417414_at 1439833_at 1444778_at | 10925 | 0.006 | 0.2004 | No | ||
| 68 | SULT2B1 | 1417335_at | 11314 | -0.003 | 0.1826 | No | ||
| 69 | MYT1 | 1422773_at 1439365_at | 11463 | -0.006 | 0.1760 | No | ||
| 70 | DOC2B | 1420666_at 1420667_at | 11555 | -0.008 | 0.1719 | No | ||
| 71 | DLL3 | 1449236_at | 11627 | -0.009 | 0.1689 | No | ||
| 72 | BMP4 | 1422912_at | 11738 | -0.012 | 0.1640 | No | ||
| 73 | HSD3B7 | 1416968_a_at 1442912_at | 11852 | -0.014 | 0.1591 | No | ||
| 74 | ESRRG | 1421747_at 1455267_at 1457896_at | 12151 | -0.020 | 0.1458 | No | ||
| 75 | FNDC5 | 1435115_at 1453135_at | 12201 | -0.021 | 0.1440 | No | ||
| 76 | PDGFB | 1450413_at 1450414_at | 12506 | -0.028 | 0.1306 | No | ||
| 77 | F11R | 1424595_at 1436374_x_at | 12640 | -0.031 | 0.1251 | No | ||
| 78 | ADAMTS2 | 1435990_at 1455720_at 1457058_at | 12660 | -0.031 | 0.1248 | No | ||
| 79 | MRPL14 | 1451307_at | 12715 | -0.032 | 0.1229 | No | ||
| 80 | MRGPRF | 1425894_at | 12842 | -0.035 | 0.1178 | No | ||
| 81 | PHOX2A | 1418620_at | 12922 | -0.037 | 0.1149 | No | ||
| 82 | USP5 | 1448311_at | 13333 | -0.046 | 0.0970 | No | ||
| 83 | ADAM15 | 1416080_at 1425170_a_at 1438760_x_at 1454206_a_at | 13579 | -0.052 | 0.0868 | No | ||
| 84 | BAZ2A | 1427209_at 1427210_at 1438192_s_at | 14054 | -0.063 | 0.0662 | No | ||
| 85 | RASL10B | 1433566_at | 14083 | -0.064 | 0.0662 | No | ||
| 86 | LZTS2 | 1426409_at | 14116 | -0.065 | 0.0659 | No | ||
| 87 | RIMS1 | 1435667_at 1438305_at 1444393_at | 14122 | -0.065 | 0.0669 | No | ||
| 88 | PSME1 | 1417056_at | 14403 | -0.073 | 0.0555 | No | ||
| 89 | JAG1 | 1421105_at 1421106_at 1434070_at | 14646 | -0.079 | 0.0459 | No | ||
| 90 | GRIN2D | 1421393_at | 14655 | -0.079 | 0.0470 | No | ||
| 91 | NOL4 | 1430189_at 1441447_at 1456387_at | 14686 | -0.080 | 0.0472 | No | ||
| 92 | IMPDH1 | 1423239_at | 14764 | -0.082 | 0.0452 | No | ||
| 93 | SIX5 | 1427560_at | 14962 | -0.088 | 0.0379 | No | ||
| 94 | MAP1A | 1460566_at | 15163 | -0.093 | 0.0305 | No | ||
| 95 | PTK7 | 1452589_at | 15252 | -0.096 | 0.0283 | No | ||
| 96 | LOXL4 | 1421153_at 1450134_at | 15373 | -0.099 | 0.0247 | No | ||
| 97 | APLN | 1451038_at | 15887 | -0.115 | 0.0033 | No | ||
| 98 | SLC22A17 | 1448209_a_at | 16114 | -0.123 | -0.0047 | No | ||
| 99 | HYAL2 | 1448679_at | 16297 | -0.129 | -0.0106 | No | ||
| 100 | VAMP1 | 1421862_a_at 1421863_at | 16461 | -0.134 | -0.0155 | No | ||
| 101 | HOXB8 | 1452493_s_at | 16753 | -0.146 | -0.0261 | No | ||
| 102 | OLFML2A | 1441635_at 1455743_at | 17346 | -0.173 | -0.0499 | No | ||
| 103 | PRKACA | 1447720_x_at 1450519_a_at | 17392 | -0.175 | -0.0486 | No | ||
| 104 | CALM2 | 1422414_a_at 1423807_a_at | 17572 | -0.187 | -0.0533 | No | ||
| 105 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 17892 | -0.208 | -0.0640 | No | ||
| 106 | BAHD1 | 1433703_s_at | 17978 | -0.213 | -0.0638 | No | ||
| 107 | OSBPL10 | 1424870_at 1442713_at 1456146_at | 18204 | -0.230 | -0.0697 | No | ||
| 108 | KCNN2 | 1445676_at 1448927_at | 18385 | -0.245 | -0.0733 | No | ||
| 109 | RBX1 | 1416577_a_at 1416578_at 1438623_x_at 1440061_at 1447793_x_at 1448387_at | 18440 | -0.249 | -0.0711 | No | ||
| 110 | VPS45A | 1418143_at | 18466 | -0.252 | -0.0674 | No | ||
| 111 | SMARCA5 | 1440048_at | 18481 | -0.253 | -0.0632 | No | ||
| 112 | PLXNC1 | 1423213_at 1423214_at 1443433_at 1443434_s_at 1450905_at 1450906_at | 18628 | -0.267 | -0.0649 | No | ||
| 113 | RELA | 1419536_a_at | 18783 | -0.282 | -0.0665 | No | ||
| 114 | SPAG6 | 1417528_at | 18925 | -0.295 | -0.0674 | No | ||
| 115 | MGAT1 | 1423609_a_at | 19169 | -0.327 | -0.0723 | No | ||
| 116 | KCNMA1 | 1424848_at 1425987_a_at 1440728_at 1444973_at 1445774_at 1457018_at | 19481 | -0.372 | -0.0795 | No | ||
| 117 | KRTCAP2 | 1417058_a_at 1417059_at | 19654 | -0.398 | -0.0798 | No | ||
| 118 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 19734 | -0.411 | -0.0756 | No | ||
| 119 | CBX6 | 1424407_s_at 1429290_at | 19904 | -0.447 | -0.0748 | No | ||
| 120 | BCL2L2 | 1423572_at 1430453_a_at 1430454_x_at 1451029_at | 20053 | -0.483 | -0.0724 | No | ||
| 121 | TRERF1 | 1444654_at | 20236 | -0.533 | -0.0706 | No | ||
| 122 | CDC42EP3 | 1422642_at 1450700_at | 20302 | -0.553 | -0.0630 | No | ||
| 123 | GALNT10 | 1418194_at 1418195_at 1440493_at | 20304 | -0.553 | -0.0525 | No | ||
| 124 | CDK2 | 1416873_a_at 1447617_at | 20384 | -0.575 | -0.0452 | No | ||
| 125 | ABCB9 | 1416263_at 1416264_at 1430394_a_at 1443636_at | 20391 | -0.576 | -0.0345 | No | ||
| 126 | NDUFA4 | 1424085_at | 20427 | -0.589 | -0.0249 | No | ||
| 127 | CAPN5 | 1418671_at 1449165_at | 20555 | -0.635 | -0.0186 | No | ||
| 128 | ABTB2 | 1433453_a_at 1433454_at 1443497_at 1444771_at | 21610 | -1.790 | -0.0328 | No | ||
| 129 | PRICKLE1 | 1441663_at 1442172_at 1442400_at 1444759_at 1452249_at 1457637_at | 21775 | -2.507 | 0.0074 | No |