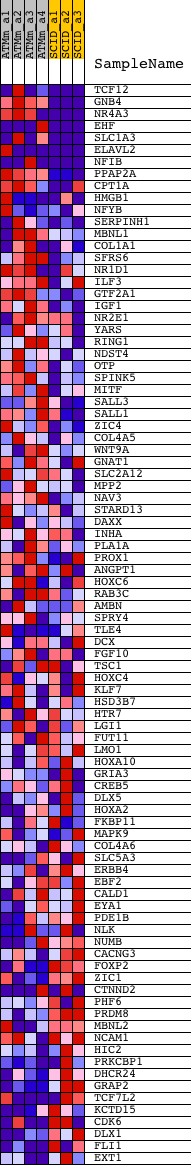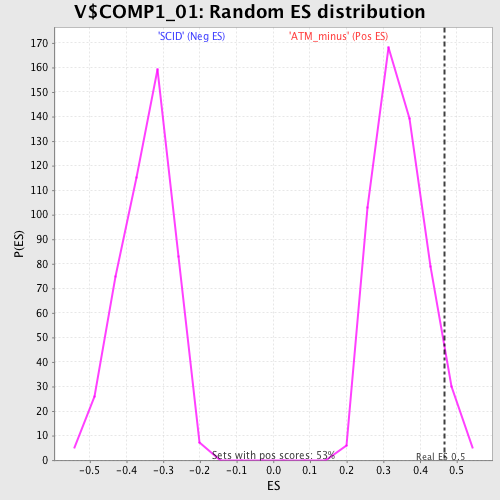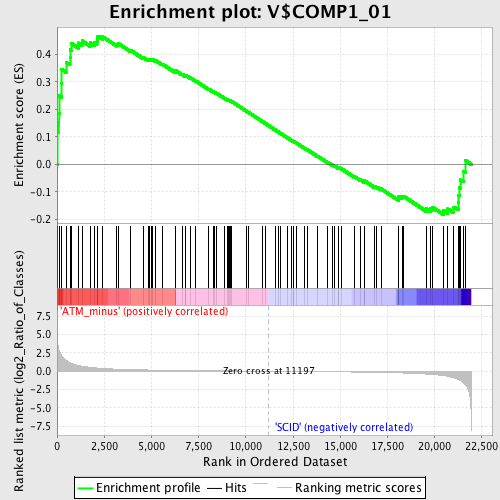
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | ATM_minus |
| GeneSet | V$COMP1_01 |
| Enrichment Score (ES) | 0.46636996 |
| Normalized Enrichment Score (NES) | 1.3458015 |
| Nominal p-value | 0.050943397 |
| FDR q-value | 0.63783425 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TCF12 | 1421908_a_at 1427670_a_at 1438762_at 1439209_at 1439619_at 1443012_at 1444620_at 1445093_at 1445133_at 1457524_at 1458337_at | 11 | 4.990 | 0.1188 | Yes | ||
| 2 | GNB4 | 1419469_at 1419470_at | 99 | 2.892 | 0.1840 | Yes | ||
| 3 | NR4A3 | 1421079_at 1421080_at | 108 | 2.785 | 0.2503 | Yes | ||
| 4 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 235 | 2.127 | 0.2954 | Yes | ||
| 5 | SLC1A3 | 1426340_at 1426341_at 1439072_at 1440491_at 1443749_x_at 1452031_at | 243 | 2.099 | 0.3453 | Yes | ||
| 6 | ELAVL2 | 1421881_a_at 1421882_a_at 1421883_at 1433052_at | 474 | 1.509 | 0.3708 | Yes | ||
| 7 | NFIB | 1416293_at 1427680_a_at 1434101_at 1434102_at 1438072_at 1438244_at 1442214_at 1444800_at 1448288_at 1454834_at | 683 | 1.164 | 0.3891 | Yes | ||
| 8 | PPAP2A | 1422619_at 1422620_s_at 1425449_at | 689 | 1.154 | 0.4165 | Yes | ||
| 9 | CPT1A | 1434866_x_at 1438156_x_at 1460409_at | 743 | 1.097 | 0.4403 | Yes | ||
| 10 | HMGB1 | 1416176_at 1425048_a_at 1435324_x_at | 1132 | 0.788 | 0.4414 | Yes | ||
| 11 | NFYB | 1419266_at 1419267_at | 1321 | 0.682 | 0.4491 | Yes | ||
| 12 | SERPINH1 | 1450843_a_at 1456733_x_at | 1747 | 0.539 | 0.4425 | Yes | ||
| 13 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 1999 | 0.471 | 0.4423 | Yes | ||
| 14 | COL1A1 | 1423669_at 1455494_at | 2113 | 0.441 | 0.4477 | Yes | ||
| 15 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 2139 | 0.436 | 0.4570 | Yes | ||
| 16 | NR1D1 | 1421802_at 1426464_at | 2160 | 0.431 | 0.4664 | Yes | ||
| 17 | ILF3 | 1422546_at 1436802_at 1460669_at | 2425 | 0.380 | 0.4634 | No | ||
| 18 | GTF2A1 | 1421357_at | 3150 | 0.282 | 0.4370 | No | ||
| 19 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 3229 | 0.274 | 0.4400 | No | ||
| 20 | NR2E1 | 1434921_at 1457289_at | 3892 | 0.219 | 0.4149 | No | ||
| 21 | YARS | 1460638_at | 4562 | 0.179 | 0.3886 | No | ||
| 22 | RING1 | 1422647_at | 4827 | 0.166 | 0.3804 | No | ||
| 23 | NDST4 | 1421318_at 1431452_at 1450195_at | 4915 | 0.162 | 0.3803 | No | ||
| 24 | OTP | 1422285_at | 4982 | 0.159 | 0.3811 | No | ||
| 25 | SPINK5 | 1430567_at | 5058 | 0.155 | 0.3814 | No | ||
| 26 | MITF | 1422025_at 1455214_at | 5203 | 0.150 | 0.3784 | No | ||
| 27 | SALL3 | 1441186_at 1452386_at | 5573 | 0.135 | 0.3647 | No | ||
| 28 | SALL1 | 1450489_at | 6246 | 0.113 | 0.3367 | No | ||
| 29 | ZIC4 | 1421539_at 1456417_at | 6275 | 0.113 | 0.3381 | No | ||
| 30 | COL4A5 | 1425475_at 1425476_at | 6281 | 0.112 | 0.3405 | No | ||
| 31 | WNT9A | 1425889_at 1436978_at | 6622 | 0.102 | 0.3274 | No | ||
| 32 | GNAT1 | 1460212_at | 6795 | 0.098 | 0.3219 | No | ||
| 33 | SLC2A12 | 1435911_s_at 1455501_at | 6799 | 0.097 | 0.3241 | No | ||
| 34 | MPP2 | 1449173_at | 7062 | 0.090 | 0.3143 | No | ||
| 35 | NAV3 | 1459440_at | 7352 | 0.083 | 0.3030 | No | ||
| 36 | STARD13 | 1452604_at | 8041 | 0.066 | 0.2731 | No | ||
| 37 | DAXX | 1419026_at | 8266 | 0.062 | 0.2643 | No | ||
| 38 | INHA | 1422728_at | 8349 | 0.059 | 0.2620 | No | ||
| 39 | PLA1A | 1417785_at | 8462 | 0.057 | 0.2582 | No | ||
| 40 | PROX1 | 1421336_at 1437894_at 1457432_at | 8878 | 0.047 | 0.2403 | No | ||
| 41 | ANGPT1 | 1421441_at 1438936_s_at 1438937_x_at 1439066_at 1446656_at 1450717_at 1457381_at | 9034 | 0.043 | 0.2343 | No | ||
| 42 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 9075 | 0.043 | 0.2335 | No | ||
| 43 | RAB3C | 1432415_at 1432432_a_at 1449494_at | 9129 | 0.041 | 0.2320 | No | ||
| 44 | AMBN | 1449417_at | 9195 | 0.040 | 0.2300 | No | ||
| 45 | SPRY4 | 1422020_at 1422021_at 1440867_at 1445669_at | 9216 | 0.040 | 0.2301 | No | ||
| 46 | TLE4 | 1425650_at 1430384_at 1450853_at | 10054 | 0.022 | 0.1923 | No | ||
| 47 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 10123 | 0.021 | 0.1897 | No | ||
| 48 | FGF10 | 1420690_at | 10893 | 0.006 | 0.1546 | No | ||
| 49 | TSC1 | 1422043_at 1439989_at 1455252_at | 11052 | 0.003 | 0.1475 | No | ||
| 50 | HOXC4 | 1422870_at | 11549 | -0.008 | 0.1250 | No | ||
| 51 | KLF7 | 1419354_at 1419355_at 1419356_at 1449439_at 1458389_at | 11710 | -0.011 | 0.1179 | No | ||
| 52 | HSD3B7 | 1416968_a_at 1442912_at | 11852 | -0.014 | 0.1118 | No | ||
| 53 | HTR7 | 1422235_at | 12218 | -0.022 | 0.0956 | No | ||
| 54 | LGI1 | 1423183_at 1435851_at | 12402 | -0.026 | 0.0878 | No | ||
| 55 | FUT11 | 1431066_at 1434123_at 1437748_at | 12525 | -0.028 | 0.0829 | No | ||
| 56 | LMO1 | 1418478_at | 12533 | -0.028 | 0.0833 | No | ||
| 57 | HOXA10 | 1431475_a_at 1446408_at | 12703 | -0.032 | 0.0763 | No | ||
| 58 | GRIA3 | 1420563_at 1434728_at | 13110 | -0.041 | 0.0587 | No | ||
| 59 | CREB5 | 1440089_at 1442576_at 1457222_at | 13244 | -0.044 | 0.0537 | No | ||
| 60 | DLX5 | 1449863_a_at | 13816 | -0.057 | 0.0289 | No | ||
| 61 | HOXA2 | 1419602_at | 14336 | -0.071 | 0.0069 | No | ||
| 62 | FKBP11 | 1417267_s_at 1425839_at 1425938_a_at | 14578 | -0.077 | -0.0023 | No | ||
| 63 | MAPK9 | 1421876_at 1421877_at 1421878_at | 14712 | -0.081 | -0.0065 | No | ||
| 64 | COL4A6 | 1421006_at 1421007_at | 14877 | -0.085 | -0.0119 | No | ||
| 65 | SLC5A3 | 1422170_at | 14899 | -0.086 | -0.0108 | No | ||
| 66 | ERBB4 | 1427783_at 1446211_at 1447043_at | 15046 | -0.090 | -0.0154 | No | ||
| 67 | EBF2 | 1418494_at 1446128_at 1449101_at 1449102_at | 15763 | -0.111 | -0.0455 | No | ||
| 68 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 16059 | -0.121 | -0.0561 | No | ||
| 69 | EYA1 | 1421727_at 1443117_at 1457424_at | 16261 | -0.128 | -0.0622 | No | ||
| 70 | PDE1B | 1449420_at | 16263 | -0.128 | -0.0592 | No | ||
| 71 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 16818 | -0.149 | -0.0810 | No | ||
| 72 | NUMB | 1416891_at 1425368_a_at 1444337_at 1457175_at | 16935 | -0.153 | -0.0827 | No | ||
| 73 | CACNG3 | 1450520_at | 17172 | -0.164 | -0.0896 | No | ||
| 74 | FOXP2 | 1422014_at 1438231_at 1438232_at 1440108_at 1441365_at 1458191_at | 18070 | -0.220 | -0.1254 | No | ||
| 75 | ZIC1 | 1423477_at 1439627_at | 18082 | -0.221 | -0.1206 | No | ||
| 76 | CTNND2 | 1422592_at 1438891_at 1443638_at 1445114_at 1447552_s_at 1456116_at | 18093 | -0.221 | -0.1158 | No | ||
| 77 | PHF6 | 1430415_at 1453761_at 1454625_at | 18271 | -0.234 | -0.1182 | No | ||
| 78 | PRDM8 | 1431852_at 1455925_at | 18365 | -0.242 | -0.1167 | No | ||
| 79 | MBNL2 | 1433754_at 1436858_at 1446475_at 1455827_at 1457237_at 1457477_at | 19548 | -0.382 | -0.1617 | No | ||
| 80 | NCAM1 | 1421966_at 1425126_at 1426864_a_at 1426865_a_at 1439556_at 1440067_at 1441995_at 1442680_at 1443018_at 1450437_a_at 1450438_at 1454140_at | 19767 | -0.421 | -0.1616 | No | ||
| 81 | HIC2 | 1427581_at 1437571_at 1454944_at | 19877 | -0.441 | -0.1560 | No | ||
| 82 | PRKCBP1 | 1426614_at 1429415_at 1445827_at 1456143_at 1458617_at | 20457 | -0.603 | -0.1681 | No | ||
| 83 | DHCR24 | 1418129_at 1451895_a_at | 20699 | -0.699 | -0.1624 | No | ||
| 84 | GRAP2 | 1456432_at | 21016 | -0.901 | -0.1553 | No | ||
| 85 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 21245 | -1.128 | -0.1388 | No | ||
| 86 | KCTD15 | 1435339_at | 21273 | -1.161 | -0.1123 | No | ||
| 87 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 21301 | -1.204 | -0.0847 | No | ||
| 88 | DLX1 | 1449470_at | 21371 | -1.298 | -0.0569 | No | ||
| 89 | FLI1 | 1422024_at 1433512_at 1441584_at | 21536 | -1.610 | -0.0259 | No | ||
| 90 | EXT1 | 1417730_at 1439877_at 1440011_at 1440092_at 1456655_at 1458054_at 1458296_at | 21631 | -1.845 | 0.0140 | No |