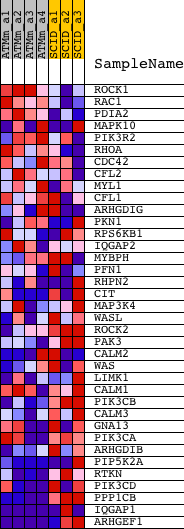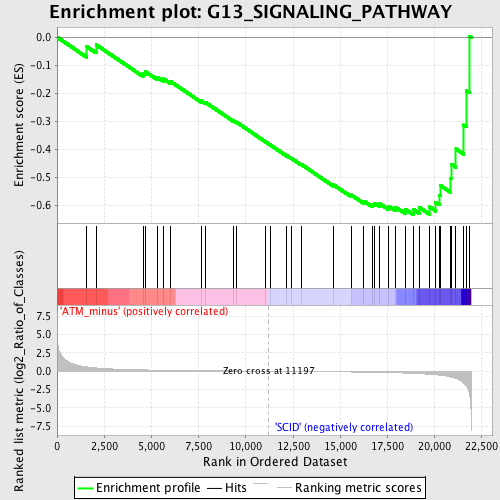
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_SCID.phenotype_ATM_minus_versus_SCID.cls #ATM_minus_versus_SCID |
| Phenotype | phenotype_ATM_minus_versus_SCID.cls#ATM_minus_versus_SCID |
| Upregulated in class | SCID |
| GeneSet | G13_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.6323736 |
| Normalized Enrichment Score (NES) | -1.5862678 |
| Nominal p-value | 0.016032064 |
| FDR q-value | 0.7772825 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 1574 | 0.592 | -0.0335 | No | ||
| 2 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 2072 | 0.450 | -0.0270 | No | ||
| 3 | PDIA2 | 1428952_at | 4551 | 0.179 | -0.1285 | No | ||
| 4 | MAPK10 | 1437195_x_at 1448342_at 1458002_at 1458062_at 1458513_at | 4684 | 0.173 | -0.1234 | No | ||
| 5 | PIK3R2 | 1418463_at | 5301 | 0.146 | -0.1421 | No | ||
| 6 | RHOA | 1437628_s_at | 5643 | 0.133 | -0.1490 | No | ||
| 7 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 6018 | 0.121 | -0.1583 | No | ||
| 8 | CFL2 | 1418066_at 1418067_at 1431432_at | 7631 | 0.076 | -0.2269 | No | ||
| 9 | MYL1 | 1452651_a_at | 7832 | 0.071 | -0.2315 | No | ||
| 10 | CFL1 | 1448346_at 1455138_x_at | 9356 | 0.037 | -0.2986 | No | ||
| 11 | ARHGDIG | 1448660_at | 9516 | 0.033 | -0.3037 | No | ||
| 12 | PKN1 | 1458674_at 1459231_at | 11011 | 0.004 | -0.3717 | No | ||
| 13 | RPS6KB1 | 1428849_at 1446376_at 1454956_at 1457562_at 1459951_at 1460705_at | 11308 | -0.003 | -0.3851 | No | ||
| 14 | IQGAP2 | 1433885_at 1459894_at | 12173 | -0.021 | -0.4232 | No | ||
| 15 | MYBPH | 1419487_at | 12400 | -0.026 | -0.4319 | No | ||
| 16 | PFN1 | 1449018_at | 12927 | -0.037 | -0.4535 | No | ||
| 17 | RHPN2 | 1431805_a_at 1434628_a_at | 14650 | -0.079 | -0.5270 | No | ||
| 18 | CIT | 1426028_a_at 1427669_a_at 1458276_x_at | 15585 | -0.105 | -0.5628 | No | ||
| 19 | MAP3K4 | 1421450_a_at 1447667_x_at 1450253_a_at 1459800_s_at | 16241 | -0.127 | -0.5844 | No | ||
| 20 | WASL | 1426776_at 1426777_a_at 1432155_at 1439832_at 1452193_a_at | 16696 | -0.144 | -0.5958 | No | ||
| 21 | ROCK2 | 1423592_at 1451041_at | 16819 | -0.149 | -0.5917 | No | ||
| 22 | PAK3 | 1417923_at 1417924_at 1437318_at | 17096 | -0.160 | -0.5940 | No | ||
| 23 | CALM2 | 1422414_a_at 1423807_a_at | 17572 | -0.187 | -0.6035 | No | ||
| 24 | WAS | 1419631_at | 17917 | -0.209 | -0.6057 | No | ||
| 25 | LIMK1 | 1417627_a_at 1425836_a_at 1456234_at 1458989_at | 18445 | -0.250 | -0.6136 | No | ||
| 26 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 18858 | -0.288 | -0.6137 | Yes | ||
| 27 | PIK3CB | 1445731_at 1453069_at | 19191 | -0.330 | -0.6075 | Yes | ||
| 28 | CALM3 | 1426710_at 1438825_at 1438826_x_at 1450864_at | 19734 | -0.411 | -0.6056 | Yes | ||
| 29 | GNA13 | 1422555_s_at 1422556_at 1430295_at 1433749_at 1446722_at 1450656_at 1453470_a_at 1460317_s_at | 20057 | -0.484 | -0.5889 | Yes | ||
| 30 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 20260 | -0.543 | -0.5630 | Yes | ||
| 31 | ARHGDIB | 1426454_at | 20297 | -0.551 | -0.5289 | Yes | ||
| 32 | PIP5K2A | 1419279_at 1419280_at 1420109_at 1442310_at 1444159_at 1449404_at 1449736_at | 20847 | -0.784 | -0.5031 | Yes | ||
| 33 | RTKN | 1425601_a_at | 20876 | -0.802 | -0.4524 | Yes | ||
| 34 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 21126 | -1.005 | -0.3987 | Yes | ||
| 35 | PPP1CB | 1426205_at 1431328_at 1433540_x_at 1456462_x_at | 21533 | -1.605 | -0.3131 | Yes | ||
| 36 | IQGAP1 | 1417379_at 1417380_at 1431395_a_at 1431396_at 1434998_at 1445724_at 1458124_at | 21675 | -1.987 | -0.1908 | Yes | ||
| 37 | ARHGEF1 | 1421164_a_at 1440403_at | 21853 | -3.127 | 0.0038 | Yes |