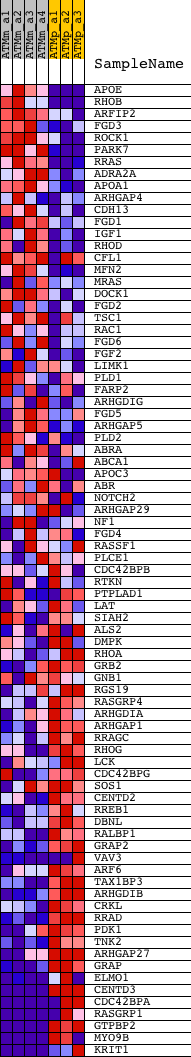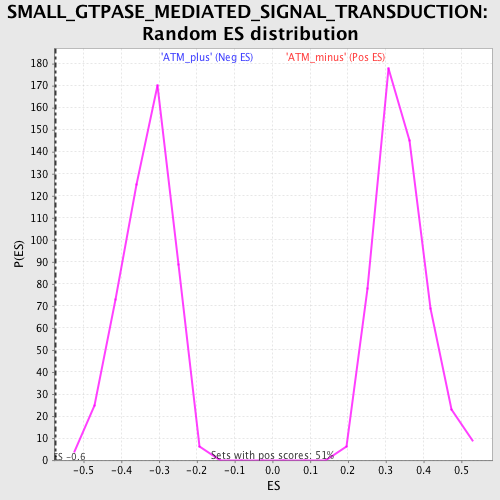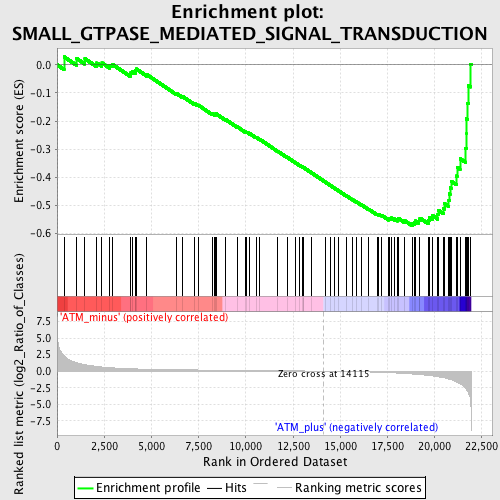
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | SMALL_GTPASE_MEDIATED_SIGNAL_TRANSDUCTION |
| Enrichment Score (ES) | -0.5728694 |
| Normalized Enrichment Score (NES) | -1.7233242 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.31049356 |
| FWER p-Value | 0.747 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | APOE | 1432466_a_at | 369 | 2.317 | 0.0284 | No | ||
| 2 | RHOB | 1449110_at | 1039 | 1.267 | 0.0225 | No | ||
| 3 | ARFIP2 | 1424240_at 1435498_at | 1473 | 0.974 | 0.0218 | No | ||
| 4 | FGD3 | 1433398_at 1450235_at 1454593_at | 2108 | 0.712 | 0.0067 | No | ||
| 5 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 2376 | 0.630 | 0.0068 | No | ||
| 6 | PARK7 | 1416526_a_at 1456194_a_at | 2781 | 0.529 | -0.0014 | No | ||
| 7 | RRAS | 1418448_at | 2917 | 0.502 | 0.0023 | No | ||
| 8 | ADRA2A | 1423022_at 1433600_at 1433601_at | 3869 | 0.368 | -0.0341 | No | ||
| 9 | APOA1 | 1419232_a_at 1419233_x_at 1438840_x_at 1455201_x_at | 3909 | 0.363 | -0.0287 | No | ||
| 10 | ARHGAP4 | 1419296_at 1443360_x_at 1458190_at | 3974 | 0.357 | -0.0247 | No | ||
| 11 | CDH13 | 1423551_at 1431824_at 1454015_a_at 1459535_at | 4136 | 0.341 | -0.0254 | No | ||
| 12 | FGD1 | 1416865_at 1437369_at 1459278_at | 4158 | 0.340 | -0.0197 | No | ||
| 13 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 4190 | 0.338 | -0.0145 | No | ||
| 14 | RHOD | 1419061_at | 4751 | 0.296 | -0.0343 | No | ||
| 15 | CFL1 | 1448346_at 1455138_x_at | 6302 | 0.224 | -0.1009 | No | ||
| 16 | MFN2 | 1441424_at 1448131_at 1451900_at | 6624 | 0.211 | -0.1115 | No | ||
| 17 | MRAS | 1449590_a_at | 7283 | 0.186 | -0.1379 | No | ||
| 18 | DOCK1 | 1443390_at 1443760_at 1443991_at 1452220_at 1457806_at | 7467 | 0.180 | -0.1428 | No | ||
| 19 | FGD2 | 1419515_at | 8220 | 0.158 | -0.1741 | No | ||
| 20 | TSC1 | 1422043_at 1439989_at 1455252_at | 8344 | 0.154 | -0.1767 | No | ||
| 21 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 8362 | 0.154 | -0.1745 | No | ||
| 22 | FGD6 | 1419322_at 1435467_at | 8416 | 0.152 | -0.1739 | No | ||
| 23 | FGF2 | 1449826_a_at | 8924 | 0.137 | -0.1945 | No | ||
| 24 | LIMK1 | 1417627_a_at 1425836_a_at 1456234_at 1458989_at | 9540 | 0.121 | -0.2202 | No | ||
| 25 | PLD1 | 1425739_at 1437112_at 1437113_s_at | 9968 | 0.109 | -0.2376 | No | ||
| 26 | FARP2 | 1424583_at 1435985_at 1439681_at 1440799_s_at | 10056 | 0.107 | -0.2395 | No | ||
| 27 | ARHGDIG | 1448660_at | 10211 | 0.103 | -0.2446 | No | ||
| 28 | FGD5 | 1460578_at | 10545 | 0.095 | -0.2580 | No | ||
| 29 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 10717 | 0.091 | -0.2640 | No | ||
| 30 | PLD2 | 1417237_at 1457252_x_at | 11688 | 0.066 | -0.3071 | No | ||
| 31 | ABRA | 1458455_at | 12211 | 0.053 | -0.3299 | No | ||
| 32 | ABCA1 | 1421839_at 1421840_at 1450392_at | 12616 | 0.043 | -0.3476 | No | ||
| 33 | APOC3 | 1418278_at | 12848 | 0.037 | -0.3574 | No | ||
| 34 | ABR | 1433477_at | 12976 | 0.033 | -0.3626 | No | ||
| 35 | NOTCH2 | 1451889_at 1455556_at | 13039 | 0.032 | -0.3648 | No | ||
| 36 | ARHGAP29 | 1441618_at 1444512_at 1454745_at | 13061 | 0.031 | -0.3652 | No | ||
| 37 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 13461 | 0.020 | -0.3830 | No | ||
| 38 | FGD4 | 1425037_at 1426041_a_at 1426042_at 1451659_at 1455337_at | 14222 | -0.004 | -0.4177 | No | ||
| 39 | RASSF1 | 1441737_s_at 1448855_at 1456994_at 1457788_at | 14460 | -0.012 | -0.4283 | No | ||
| 40 | PLCE1 | 1452398_at | 14701 | -0.021 | -0.4389 | No | ||
| 41 | CDC42BPB | 1434652_at 1459167_at | 14894 | -0.029 | -0.4471 | No | ||
| 42 | RTKN | 1425601_a_at | 15351 | -0.047 | -0.4671 | No | ||
| 43 | PTPLAD1 | 1433706_a_at 1452427_s_at 1454698_at | 15650 | -0.062 | -0.4795 | No | ||
| 44 | LAT | 1460651_at | 15852 | -0.072 | -0.4873 | No | ||
| 45 | SIAH2 | 1448170_at 1448171_at | 16119 | -0.088 | -0.4977 | No | ||
| 46 | ALS2 | 1417783_at 1417784_at 1431608_at 1446183_at | 16495 | -0.112 | -0.5127 | No | ||
| 47 | DMPK | 1422989_a_at 1434944_at | 16950 | -0.149 | -0.5305 | No | ||
| 48 | RHOA | 1437628_s_at | 17032 | -0.156 | -0.5312 | No | ||
| 49 | GRB2 | 1418508_a_at 1449111_a_at | 17171 | -0.170 | -0.5342 | No | ||
| 50 | GNB1 | 1417432_a_at 1425908_at 1454696_at 1459442_at | 17559 | -0.220 | -0.5476 | No | ||
| 51 | RGS19 | 1417786_a_at 1434940_x_at | 17612 | -0.228 | -0.5455 | No | ||
| 52 | RASGRP4 | 1425380_at | 17686 | -0.238 | -0.5442 | No | ||
| 53 | ARHGDIA | 1451168_a_at | 17877 | -0.262 | -0.5478 | No | ||
| 54 | ARHGAP1 | 1424307_at 1451309_at | 18044 | -0.284 | -0.5498 | No | ||
| 55 | RRAGC | 1415749_a_at | 18063 | -0.287 | -0.5450 | No | ||
| 56 | RHOG | 1422572_at | 18397 | -0.339 | -0.5537 | No | ||
| 57 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 18818 | -0.421 | -0.5646 | Yes | ||
| 58 | CDC42BPG | 1436197_at | 18917 | -0.444 | -0.5604 | Yes | ||
| 59 | SOS1 | 1421884_at 1421885_at 1421886_at | 18996 | -0.463 | -0.5550 | Yes | ||
| 60 | CENTD2 | 1428064_at | 19167 | -0.504 | -0.5529 | Yes | ||
| 61 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 19216 | -0.514 | -0.5450 | Yes | ||
| 62 | DBNL | 1460334_at | 19671 | -0.646 | -0.5532 | Yes | ||
| 63 | RALBP1 | 1417248_at 1448634_at | 19716 | -0.659 | -0.5423 | Yes | ||
| 64 | GRAP2 | 1456432_at | 19875 | -0.712 | -0.5356 | Yes | ||
| 65 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20163 | -0.833 | -0.5325 | Yes | ||
| 66 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 20186 | -0.844 | -0.5170 | Yes | ||
| 67 | TAX1BP3 | 1424169_at | 20455 | -0.973 | -0.5102 | Yes | ||
| 68 | ARHGDIB | 1426454_at | 20514 | -1.004 | -0.4933 | Yes | ||
| 69 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 20749 | -1.171 | -0.4811 | Yes | ||
| 70 | RRAD | 1422562_at | 20772 | -1.195 | -0.4587 | Yes | ||
| 71 | PDK1 | 1423747_a_at 1423748_at 1434974_at 1435836_at | 20815 | -1.229 | -0.4366 | Yes | ||
| 72 | TNK2 | 1448298_at | 20872 | -1.276 | -0.4143 | Yes | ||
| 73 | ARHGAP27 | 1416758_at 1457655_x_at | 21152 | -1.636 | -0.3951 | Yes | ||
| 74 | GRAP | 1429387_at | 21185 | -1.677 | -0.3637 | Yes | ||
| 75 | ELMO1 | 1424523_at 1446610_at 1450208_a_at | 21343 | -1.914 | -0.3335 | Yes | ||
| 76 | CENTD3 | 1419833_s_at 1447407_at 1451282_at | 21634 | -2.598 | -0.2960 | Yes | ||
| 77 | CDC42BPA | 1435847_at 1436380_at 1438449_at 1442260_at 1446945_at | 21666 | -2.698 | -0.2447 | Yes | ||
| 78 | RASGRP1 | 1421176_at 1431749_a_at 1434295_at 1450143_at | 21672 | -2.736 | -0.1915 | Yes | ||
| 79 | GTPBP2 | 1416690_at 1416691_at 1442305_at 1448437_a_at | 21748 | -3.015 | -0.1360 | Yes | ||
| 80 | MYO9B | 1418031_at 1438533_at 1459110_at | 21776 | -3.233 | -0.0740 | Yes | ||
| 81 | KRIT1 | 1428729_at 1428730_at 1439317_at 1442125_at 1448701_a_at | 21882 | -4.161 | 0.0025 | Yes |