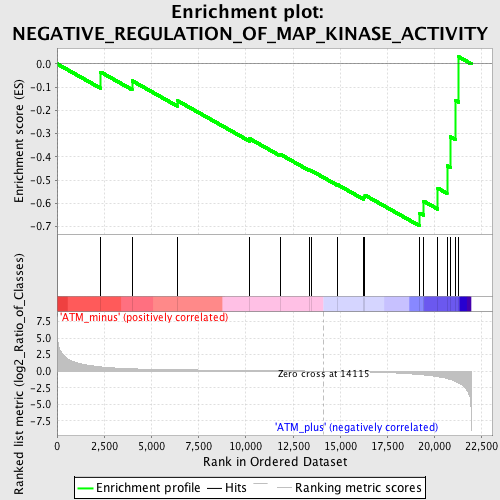
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NEGATIVE_REGULATION_OF_MAP_KINASE_ACTIVITY |
| Enrichment Score (ES) | -0.6969787 |
| Normalized Enrichment Score (NES) | -1.5440385 |
| Nominal p-value | 0.020703934 |
| FDR q-value | 0.63254786 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DUSP9 | 1433844_a_at 1433845_x_at 1454737_at | 2304 | 0.650 | -0.0351 | No | ||
| 2 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 3972 | 0.357 | -0.0726 | No | ||
| 3 | DUSP8 | 1418714_at | 6361 | 0.221 | -0.1577 | No | ||
| 4 | RGS4 | 1416286_at 1416287_at 1448285_at | 10194 | 0.103 | -0.3214 | No | ||
| 5 | SPRED1 | 1423160_at 1423161_s_at 1423162_s_at 1428777_at 1441517_at 1442859_at 1443652_x_at 1445228_at 1452911_at 1460116_s_at | 11822 | 0.062 | -0.3889 | No | ||
| 6 | SPRED2 | 1423001_at 1434403_at 1436892_at 1441415_at 1441802_at | 13352 | 0.023 | -0.4562 | No | ||
| 7 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 13461 | 0.020 | -0.4590 | No | ||
| 8 | GPS2 | 1417796_at | 14873 | -0.028 | -0.5203 | No | ||
| 9 | MBIP | 1424405_at | 16243 | -0.095 | -0.5725 | No | ||
| 10 | GPS1 | 1415699_a_at | 16305 | -0.100 | -0.5645 | No | ||
| 11 | PDCD4 | 1444171_at | 19209 | -0.513 | -0.6417 | Yes | ||
| 12 | DUSP22 | 1441723_at 1448985_at | 19407 | -0.562 | -0.5901 | Yes | ||
| 13 | LAX1 | 1438687_at | 20166 | -0.836 | -0.5347 | Yes | ||
| 14 | DUSP16 | 1418401_a_at 1440615_at 1440651_at | 20653 | -1.097 | -0.4387 | Yes | ||
| 15 | DUSP2 | 1450698_at | 20824 | -1.233 | -0.3136 | Yes | ||
| 16 | HIPK3 | 1419191_at 1426819_at | 21115 | -1.567 | -0.1580 | Yes | ||
| 17 | DUSP6 | 1415834_at | 21275 | -1.814 | 0.0302 | Yes |

