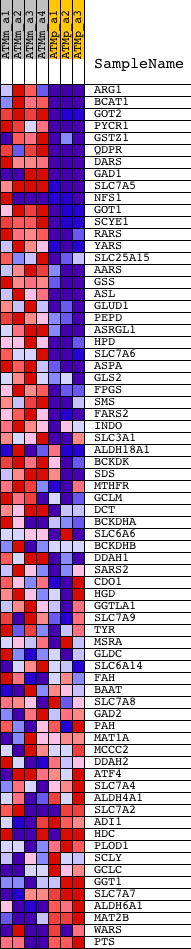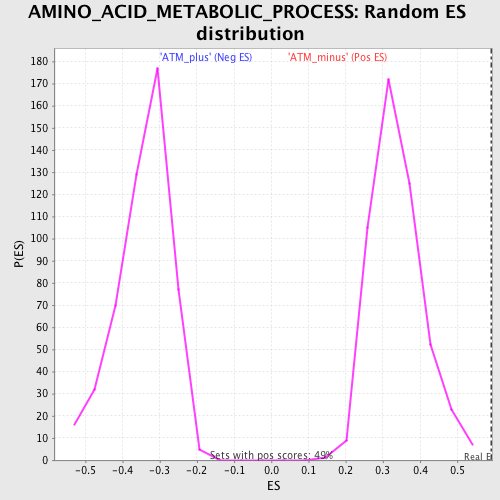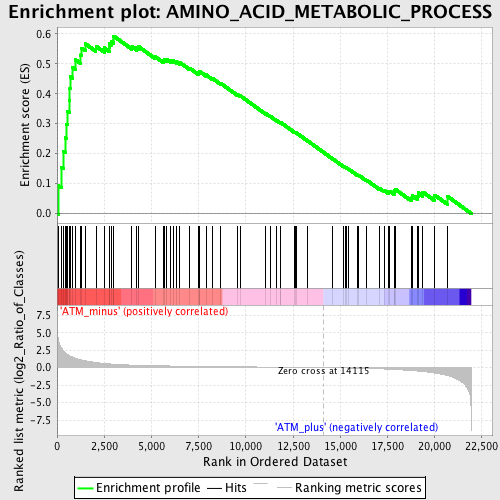
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | AMINO_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5925082 |
| Normalized Enrichment Score (NES) | 1.7572803 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.24036607 |
| FWER p-Value | 0.476 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARG1 | 1419549_at | 67 | 3.938 | 0.0926 | Yes | ||
| 2 | BCAT1 | 1430111_a_at 1450871_a_at | 220 | 2.860 | 0.1551 | Yes | ||
| 3 | GOT2 | 1417715_a_at 1417716_at | 348 | 2.373 | 0.2069 | Yes | ||
| 4 | PYCR1 | 1424556_at | 449 | 2.082 | 0.2529 | Yes | ||
| 5 | GSTZ1 | 1427552_a_at 1458134_at | 506 | 1.961 | 0.2979 | Yes | ||
| 6 | QDPR | 1423664_at 1437993_x_at | 551 | 1.872 | 0.3413 | Yes | ||
| 7 | DARS | 1423800_at | 650 | 1.705 | 0.3783 | Yes | ||
| 8 | GAD1 | 1416561_at 1416562_at | 676 | 1.672 | 0.4177 | Yes | ||
| 9 | SLC7A5 | 1418326_at | 688 | 1.654 | 0.4574 | Yes | ||
| 10 | NFS1 | 1416373_at 1431431_a_at 1442017_at | 803 | 1.517 | 0.4890 | Yes | ||
| 11 | GOT1 | 1450970_at | 968 | 1.333 | 0.5139 | Yes | ||
| 12 | SCYE1 | 1416486_at | 1226 | 1.129 | 0.5295 | Yes | ||
| 13 | RARS | 1416312_at | 1297 | 1.083 | 0.5526 | Yes | ||
| 14 | YARS | 1460638_at | 1507 | 0.963 | 0.5664 | Yes | ||
| 15 | SLC25A15 | 1420966_at 1420967_at | 2059 | 0.726 | 0.5589 | Yes | ||
| 16 | AARS | 1423685_at 1451083_s_at | 2497 | 0.595 | 0.5533 | Yes | ||
| 17 | GSS | 1441931_x_at 1448273_at | 2761 | 0.533 | 0.5543 | Yes | ||
| 18 | ASL | 1448350_at | 2767 | 0.532 | 0.5670 | Yes | ||
| 19 | GLUD1 | 1416209_at 1448253_at | 2861 | 0.515 | 0.5752 | Yes | ||
| 20 | PEPD | 1416712_at | 3002 | 0.488 | 0.5807 | Yes | ||
| 21 | ASRGL1 | 1424395_at 1424396_a_at 1460360_at | 3003 | 0.488 | 0.5925 | Yes | ||
| 22 | HPD | 1424618_at | 3964 | 0.358 | 0.5573 | No | ||
| 23 | SLC7A6 | 1433467_at 1460541_at | 4223 | 0.335 | 0.5536 | No | ||
| 24 | ASPA | 1418472_at | 4331 | 0.328 | 0.5567 | No | ||
| 25 | GLS2 | 1435245_at | 5210 | 0.272 | 0.5232 | No | ||
| 26 | FPGS | 1460673_at | 5621 | 0.253 | 0.5105 | No | ||
| 27 | SMS | 1428699_at 1434190_at | 5689 | 0.249 | 0.5135 | No | ||
| 28 | FARS2 | 1431354_a_at 1439406_x_at | 5791 | 0.245 | 0.5149 | No | ||
| 29 | INDO | 1420437_at | 6002 | 0.235 | 0.5110 | No | ||
| 30 | SLC3A1 | 1448741_at | 6146 | 0.229 | 0.5100 | No | ||
| 31 | ALDH18A1 | 1415836_at 1435855_x_at 1437325_x_at 1437333_x_at 1437620_x_at | 6332 | 0.222 | 0.5069 | No | ||
| 32 | BCKDK | 1443813_x_at 1460644_at | 6504 | 0.216 | 0.5043 | No | ||
| 33 | SDS | 1424744_at | 7036 | 0.195 | 0.4848 | No | ||
| 34 | MTHFR | 1422132_at 1434087_at 1450498_at | 7502 | 0.179 | 0.4679 | No | ||
| 35 | GCLM | 1418627_at | 7529 | 0.178 | 0.4710 | No | ||
| 36 | DCT | 1418028_at | 7553 | 0.177 | 0.4743 | No | ||
| 37 | BCKDHA | 1416647_at | 7889 | 0.168 | 0.4630 | No | ||
| 38 | SLC6A6 | 1420148_at 1421346_a_at 1437149_at 1449751_at | 8254 | 0.157 | 0.4502 | No | ||
| 39 | BCKDHB | 1427153_at | 8679 | 0.144 | 0.4343 | No | ||
| 40 | DDAH1 | 1429298_at 1429299_at 1434519_at 1438879_at 1454995_at 1455400_at | 9570 | 0.120 | 0.3965 | No | ||
| 41 | SARS2 | 1448522_at | 9700 | 0.116 | 0.3934 | No | ||
| 42 | CDO1 | 1448842_at | 11049 | 0.082 | 0.3338 | No | ||
| 43 | HGD | 1452986_at | 11284 | 0.076 | 0.3249 | No | ||
| 44 | GGTLA1 | 1418216_at 1439420_x_at 1455747_at | 11635 | 0.067 | 0.3105 | No | ||
| 45 | SLC7A9 | 1448783_at | 11821 | 0.063 | 0.3036 | No | ||
| 46 | TYR | 1417717_a_at 1448821_at 1456095_at | 11840 | 0.062 | 0.3042 | No | ||
| 47 | MSRA | 1442385_at 1448856_a_at 1458923_at | 12566 | 0.044 | 0.2722 | No | ||
| 48 | GLDC | 1416049_at 1459959_at | 12602 | 0.043 | 0.2716 | No | ||
| 49 | SLC6A14 | 1420503_at 1420504_at | 12671 | 0.042 | 0.2695 | No | ||
| 50 | FAH | 1417220_at | 13255 | 0.026 | 0.2435 | No | ||
| 51 | BAAT | 1419001_at 1419002_s_at | 14566 | -0.017 | 0.1840 | No | ||
| 52 | SLC7A8 | 1417929_at | 15143 | -0.039 | 0.1586 | No | ||
| 53 | GAD2 | 1421978_at | 15281 | -0.044 | 0.1534 | No | ||
| 54 | PAH | 1454638_a_at | 15338 | -0.047 | 0.1520 | No | ||
| 55 | MAT1A | 1423147_at | 15450 | -0.052 | 0.1481 | No | ||
| 56 | MCCC2 | 1428021_at 1432472_a_at 1454840_at | 15912 | -0.076 | 0.1289 | No | ||
| 57 | DDAH2 | 1416457_at | 15976 | -0.079 | 0.1279 | No | ||
| 58 | ATF4 | 1438992_x_at 1439258_at 1448135_at | 16381 | -0.105 | 0.1120 | No | ||
| 59 | SLC7A4 | 1426068_at 1426069_s_at 1436776_x_at | 17079 | -0.162 | 0.0841 | No | ||
| 60 | ALDH4A1 | 1452375_at | 17329 | -0.188 | 0.0772 | No | ||
| 61 | SLC7A2 | 1422648_at 1426008_a_at 1436555_at 1440506_at 1450703_at | 17573 | -0.223 | 0.0715 | No | ||
| 62 | ADI1 | 1449076_x_at | 17615 | -0.228 | 0.0752 | No | ||
| 63 | HDC | 1451796_s_at 1454713_s_at | 17848 | -0.259 | 0.0709 | No | ||
| 64 | PLOD1 | 1416289_at 1445893_at | 17870 | -0.262 | 0.0763 | No | ||
| 65 | SCLY | 1417671_at 1441192_at | 17933 | -0.269 | 0.0800 | No | ||
| 66 | GCLC | 1424296_at | 18760 | -0.407 | 0.0521 | No | ||
| 67 | GGT1 | 1448485_at | 18803 | -0.418 | 0.0604 | No | ||
| 68 | SLC7A7 | 1417392_a_at 1447181_s_at | 19105 | -0.489 | 0.0585 | No | ||
| 69 | ALDH6A1 | 1448104_at | 19125 | -0.495 | 0.0696 | No | ||
| 70 | MAT2B | 1448196_at | 19374 | -0.553 | 0.0717 | No | ||
| 71 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 20003 | -0.763 | 0.0615 | No | ||
| 72 | PTS | 1450660_at 1458402_at | 20666 | -1.106 | 0.0581 | No |