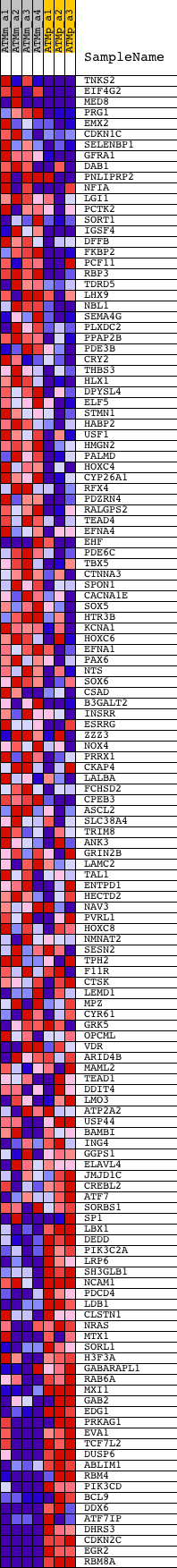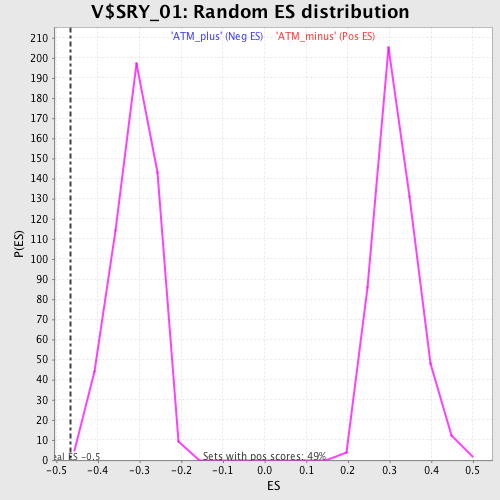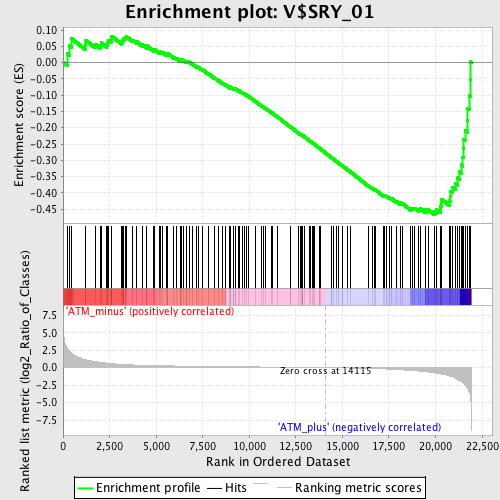
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$SRY_01 |
| Enrichment Score (ES) | -0.46639967 |
| Normalized Enrichment Score (NES) | -1.494825 |
| Nominal p-value | 0.001953125 |
| FDR q-value | 0.163362 |
| FWER p-Value | 0.857 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TNKS2 | 1428388_at 1437793_at 1447522_s_at 1452772_at 1457468_at | 217 | 2.862 | 0.0269 | No | ||
| 2 | EIF4G2 | 1415863_at 1452758_s_at | 343 | 2.384 | 0.0519 | No | ||
| 3 | MED8 | 1431423_a_at 1442417_at | 447 | 2.084 | 0.0740 | No | ||
| 4 | PRG1 | 1417426_at | 1180 | 1.158 | 0.0553 | No | ||
| 5 | EMX2 | 1456258_at | 1225 | 1.131 | 0.0679 | No | ||
| 6 | CDKN1C | 1417649_at | 1715 | 0.870 | 0.0566 | No | ||
| 7 | SELENBP1 | 1417580_s_at 1450699_at | 1989 | 0.751 | 0.0538 | No | ||
| 8 | GFRA1 | 1421973_at 1439015_at 1450440_at | 2038 | 0.735 | 0.0611 | No | ||
| 9 | DAB1 | 1421100_a_at 1427307_a_at 1427308_at 1435577_at 1435578_s_at | 2354 | 0.635 | 0.0548 | No | ||
| 10 | PNLIPRP2 | 1437438_x_at 1448186_at | 2395 | 0.624 | 0.0610 | No | ||
| 11 | NFIA | 1421162_a_at 1421163_a_at 1427733_a_at 1438236_at 1440660_at 1441547_at 1446742_at 1446990_at 1456087_at 1459431_at | 2410 | 0.621 | 0.0683 | No | ||
| 12 | LGI1 | 1423183_at 1435851_at | 2575 | 0.578 | 0.0683 | No | ||
| 13 | PCTK2 | 1435143_at 1446130_at 1446272_at | 2580 | 0.577 | 0.0755 | No | ||
| 14 | SORT1 | 1423362_at 1423363_at 1450955_s_at | 2616 | 0.569 | 0.0812 | No | ||
| 15 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 3151 | 0.461 | 0.0627 | No | ||
| 16 | DFFB | 1421229_at 1437051_at | 3182 | 0.456 | 0.0672 | No | ||
| 17 | FKBP2 | 1450694_at | 3219 | 0.448 | 0.0713 | No | ||
| 18 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 3261 | 0.442 | 0.0751 | No | ||
| 19 | RBP3 | 1425105_at 1457855_at | 3363 | 0.430 | 0.0760 | No | ||
| 20 | TDRD5 | 1446208_at 1456391_at | 3400 | 0.423 | 0.0798 | No | ||
| 21 | LHX9 | 1419324_at 1431598_a_at 1441313_x_at | 3746 | 0.382 | 0.0689 | No | ||
| 22 | NBL1 | 1448428_at | 3950 | 0.360 | 0.0642 | No | ||
| 23 | SEMA4G | 1449202_at | 4251 | 0.333 | 0.0547 | No | ||
| 24 | PLXDC2 | 1418912_at 1449270_at | 4480 | 0.314 | 0.0483 | No | ||
| 25 | PPAP2B | 1429514_at 1446850_at 1448908_at | 4484 | 0.313 | 0.0522 | No | ||
| 26 | PDE3B | 1433694_at | 4879 | 0.289 | 0.0379 | No | ||
| 27 | CRY2 | 1426383_at | 4892 | 0.288 | 0.0410 | No | ||
| 28 | THBS3 | 1416623_at | 5180 | 0.274 | 0.0314 | No | ||
| 29 | HLX1 | 1418939_at | 5229 | 0.271 | 0.0327 | No | ||
| 30 | DPYSL4 | 1418297_at 1418298_s_at 1418299_at | 5325 | 0.267 | 0.0318 | No | ||
| 31 | ELF5 | 1419555_at 1419556_at | 5569 | 0.256 | 0.0239 | No | ||
| 32 | STMN1 | 1415849_s_at | 5579 | 0.255 | 0.0268 | No | ||
| 33 | HABP2 | 1443695_at 1443696_s_at | 5630 | 0.253 | 0.0277 | No | ||
| 34 | USF1 | 1426164_a_at 1448805_at 1459758_at | 5945 | 0.238 | 0.0164 | No | ||
| 35 | HMGN2 | 1433507_a_at | 6108 | 0.230 | 0.0119 | No | ||
| 36 | PALMD | 1417251_at | 6283 | 0.224 | 0.0068 | No | ||
| 37 | HOXC4 | 1422870_at | 6294 | 0.224 | 0.0093 | No | ||
| 38 | CYP26A1 | 1419430_at | 6380 | 0.220 | 0.0082 | No | ||
| 39 | RFX4 | 1432053_at 1436931_at 1443312_at | 6469 | 0.217 | 0.0070 | No | ||
| 40 | PDZRN4 | 1456512_at | 6634 | 0.210 | 0.0021 | No | ||
| 41 | RALGPS2 | 1417230_at 1428789_at 1431704_a_at | 6641 | 0.210 | 0.0046 | No | ||
| 42 | TEAD4 | 1426337_a_at 1460683_at | 6760 | 0.205 | 0.0018 | No | ||
| 43 | EFNA4 | 1421784_a_at | 6950 | 0.198 | -0.0043 | No | ||
| 44 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 7174 | 0.191 | -0.0121 | No | ||
| 45 | PDE6C | 1450830_a_at | 7270 | 0.187 | -0.0140 | No | ||
| 46 | TBX5 | 1425694_at 1425695_at | 7506 | 0.179 | -0.0225 | No | ||
| 47 | CTNNA3 | 1441560_at | 7788 | 0.171 | -0.0332 | No | ||
| 48 | SPON1 | 1424415_s_at 1441226_at 1451342_at | 8115 | 0.161 | -0.0461 | No | ||
| 49 | CACNA1E | 1421692_at | 8358 | 0.154 | -0.0552 | No | ||
| 50 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 8574 | 0.147 | -0.0632 | No | ||
| 51 | HTR3B | 1421652_at | 8709 | 0.143 | -0.0675 | No | ||
| 52 | KCNA1 | 1417416_at 1437230_at 1442413_at 1455785_at | 8958 | 0.136 | -0.0771 | No | ||
| 53 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 8966 | 0.136 | -0.0757 | No | ||
| 54 | EFNA1 | 1416895_at 1448510_at | 8986 | 0.135 | -0.0748 | No | ||
| 55 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 9129 | 0.132 | -0.0796 | No | ||
| 56 | NTS | 1422860_at | 9164 | 0.131 | -0.0795 | No | ||
| 57 | SOX6 | 1426018_a_at 1427677_a_at 1434918_at 1447655_x_at | 9254 | 0.128 | -0.0819 | No | ||
| 58 | CSAD | 1427981_a_at | 9260 | 0.128 | -0.0805 | No | ||
| 59 | B3GALT2 | 1423084_at 1437433_at 1437644_at | 9416 | 0.124 | -0.0860 | No | ||
| 60 | INSRR | 1420564_at | 9449 | 0.123 | -0.0859 | No | ||
| 61 | ESRRG | 1421747_at 1455267_at 1457896_at | 9650 | 0.118 | -0.0936 | No | ||
| 62 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 9719 | 0.116 | -0.0952 | No | ||
| 63 | NOX4 | 1419161_a_at 1451827_a_at | 9824 | 0.113 | -0.0985 | No | ||
| 64 | PRRX1 | 1425526_a_at 1425527_at 1425528_at 1432129_a_at 1439774_at 1458004_at | 9934 | 0.110 | -0.1021 | No | ||
| 65 | CKAP4 | 1426754_x_at 1426755_at 1452181_at 1455019_x_at | 10334 | 0.100 | -0.1191 | No | ||
| 66 | LALBA | 1418363_at | 10665 | 0.092 | -0.1331 | No | ||
| 67 | FCHSD2 | 1434260_at 1456949_at | 10778 | 0.089 | -0.1370 | No | ||
| 68 | CPEB3 | 1437765_at 1445868_at 1455372_at 1456048_at | 10859 | 0.087 | -0.1396 | No | ||
| 69 | ASCL2 | 1422396_s_at 1432018_at 1460514_s_at | 11180 | 0.079 | -0.1533 | No | ||
| 70 | SLC38A4 | 1428111_at 1448889_at | 11224 | 0.078 | -0.1542 | No | ||
| 71 | TRIM8 | 1418577_at 1427593_at 1438679_at 1460237_at | 11495 | 0.071 | -0.1657 | No | ||
| 72 | ANK3 | 1425202_a_at 1439220_at 1447259_at 1451628_a_at 1457288_at | 12194 | 0.053 | -0.1970 | No | ||
| 73 | GRIN2B | 1422223_at 1431700_at | 12231 | 0.053 | -0.1980 | No | ||
| 74 | LAMC2 | 1421279_at 1452505_at | 12632 | 0.043 | -0.2158 | No | ||
| 75 | TAL1 | 1449389_at | 12741 | 0.040 | -0.2203 | No | ||
| 76 | ENTPD1 | 1423326_at 1450939_at 1453586_at | 12828 | 0.037 | -0.2237 | No | ||
| 77 | HECTD2 | 1433944_at 1438596_at 1446224_at 1446430_at | 12831 | 0.037 | -0.2233 | No | ||
| 78 | NAV3 | 1459440_at | 12849 | 0.037 | -0.2236 | No | ||
| 79 | PVRL1 | 1438111_at 1438421_at 1440131_at 1446488_at 1450819_at | 12861 | 0.036 | -0.2237 | No | ||
| 80 | HOXC8 | 1452412_at | 12969 | 0.033 | -0.2282 | No | ||
| 81 | NMNAT2 | 1436155_at | 13249 | 0.026 | -0.2406 | No | ||
| 82 | SESN2 | 1425139_at 1451599_at | 13309 | 0.024 | -0.2430 | No | ||
| 83 | TPH2 | 1435314_at | 13418 | 0.021 | -0.2477 | No | ||
| 84 | F11R | 1424595_at 1436374_x_at | 13437 | 0.021 | -0.2483 | No | ||
| 85 | CTSK | 1450652_at | 13486 | 0.019 | -0.2502 | No | ||
| 86 | LEMD1 | 1445642_at | 13761 | 0.010 | -0.2627 | No | ||
| 87 | MPZ | 1423253_at | 13786 | 0.010 | -0.2636 | No | ||
| 88 | CYR61 | 1416039_x_at 1417848_at 1417849_at 1438133_a_at 1442340_x_at 1457823_at 1459462_at | 13817 | 0.009 | -0.2649 | No | ||
| 89 | GRK5 | 1446361_at 1446998_at 1449514_at | 14402 | -0.010 | -0.2916 | No | ||
| 90 | OPCML | 1435612_at 1442752_at 1457446_at 1458228_at | 14424 | -0.011 | -0.2924 | No | ||
| 91 | VDR | 1418175_at 1418176_at | 14514 | -0.015 | -0.2963 | No | ||
| 92 | ARID4B | 1431024_a_at 1431031_at 1435768_at 1446317_at 1451847_s_at 1460384_a_at | 14682 | -0.021 | -0.3037 | No | ||
| 93 | MAML2 | 1446570_at 1447546_s_at | 14777 | -0.024 | -0.3077 | No | ||
| 94 | TEAD1 | 1423434_at 1447911_at 1456717_at | 14978 | -0.032 | -0.3164 | No | ||
| 95 | DDIT4 | 1428306_at | 14981 | -0.032 | -0.3161 | No | ||
| 96 | LMO3 | 1455754_at | 15274 | -0.044 | -0.3289 | No | ||
| 97 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 15428 | -0.051 | -0.3353 | No | ||
| 98 | USP44 | 1456690_at | 16408 | -0.106 | -0.3788 | No | ||
| 99 | BAMBI | 1423753_at | 16590 | -0.120 | -0.3856 | No | ||
| 100 | ING4 | 1448054_at 1460728_s_at | 16732 | -0.131 | -0.3904 | No | ||
| 101 | GGPS1 | 1419505_a_at 1419506_at 1419805_s_at 1429356_s_at | 16756 | -0.133 | -0.3897 | No | ||
| 102 | ELAVL4 | 1428741_at 1450258_a_at 1452894_at 1457399_at | 17201 | -0.174 | -0.4079 | No | ||
| 103 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 17234 | -0.178 | -0.4070 | No | ||
| 104 | CREBL2 | 1435085_at 1442738_at | 17357 | -0.192 | -0.4102 | No | ||
| 105 | ATF7 | 1425653_at 1437645_at 1445509_at | 17529 | -0.216 | -0.4152 | No | ||
| 106 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 17637 | -0.231 | -0.4172 | No | ||
| 107 | SP1 | 1418180_at 1448994_at 1454852_at | 17912 | -0.267 | -0.4263 | No | ||
| 108 | LBX1 | 1422323_a_at 1427400_at | 18093 | -0.292 | -0.4308 | No | ||
| 109 | DEDD | 1432000_a_at 1434994_at 1434995_s_at | 18203 | -0.309 | -0.4318 | No | ||
| 110 | PIK3C2A | 1421023_at 1425862_a_at 1443655_s_at | 18675 | -0.391 | -0.4484 | No | ||
| 111 | LRP6 | 1451022_at | 18754 | -0.405 | -0.4468 | No | ||
| 112 | SH3GLB1 | 1418010_a_at 1418011_a_at 1418012_at | 18874 | -0.435 | -0.4466 | No | ||
| 113 | NCAM1 | 1421966_at 1425126_at 1426864_a_at 1426865_a_at 1439556_at 1440067_at 1441995_at 1442680_at 1443018_at 1450437_a_at 1450438_at 1454140_at | 19097 | -0.487 | -0.4505 | No | ||
| 114 | PDCD4 | 1444171_at | 19209 | -0.513 | -0.4490 | No | ||
| 115 | LDB1 | 1452024_a_at | 19436 | -0.571 | -0.4520 | No | ||
| 116 | CLSTN1 | 1421860_at 1421861_at 1445085_at | 19596 | -0.620 | -0.4514 | No | ||
| 117 | NRAS | 1422688_a_at 1454060_a_at | 19925 | -0.733 | -0.4570 | Yes | ||
| 118 | MTX1 | 1418521_a_at 1418522_at 1447361_at | 20043 | -0.778 | -0.4523 | Yes | ||
| 119 | SORL1 | 1426258_at 1445889_at 1453003_at 1460390_at | 20248 | -0.868 | -0.4505 | Yes | ||
| 120 | H3F3A | 1423263_at 1442087_at | 20275 | -0.881 | -0.4403 | Yes | ||
| 121 | GABARAPL1 | 1416418_at 1416419_s_at 1442572_at | 20304 | -0.895 | -0.4301 | Yes | ||
| 122 | RAB6A | 1447776_x_at 1448304_a_at 1448305_at | 20335 | -0.911 | -0.4197 | Yes | ||
| 123 | MXI1 | 1425732_a_at 1450376_at | 20743 | -1.165 | -0.4234 | Yes | ||
| 124 | GAB2 | 1419829_a_at 1420785_at 1439786_at 1446860_at | 20788 | -1.204 | -0.4099 | Yes | ||
| 125 | EDG1 | 1423571_at | 20823 | -1.233 | -0.3956 | Yes | ||
| 126 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 20924 | -1.313 | -0.3833 | Yes | ||
| 127 | EVA1 | 1416236_a_at 1416237_at 1448265_x_at | 21073 | -1.510 | -0.3706 | Yes | ||
| 128 | TCF7L2 | 1425229_a_at 1426639_a_at 1429427_s_at 1429428_at 1441756_at 1446452_at 1447071_at | 21193 | -1.692 | -0.3543 | Yes | ||
| 129 | DUSP6 | 1415834_at | 21275 | -1.814 | -0.3346 | Yes | ||
| 130 | ABLIM1 | 1442376_at 1453103_at 1454708_at 1460120_at | 21392 | -1.991 | -0.3143 | Yes | ||
| 131 | RBM4 | 1416920_at 1437322_at 1455517_at | 21472 | -2.127 | -0.2905 | Yes | ||
| 132 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 21492 | -2.164 | -0.2635 | Yes | ||
| 133 | BCL9 | 1445859_at 1446767_at 1451574_at 1455565_at | 21515 | -2.210 | -0.2361 | Yes | ||
| 134 | DDX6 | 1424598_at 1457618_at | 21586 | -2.409 | -0.2083 | Yes | ||
| 135 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 21698 | -2.826 | -0.1770 | Yes | ||
| 136 | DHRS3 | 1448390_a_at | 21716 | -2.870 | -0.1408 | Yes | ||
| 137 | CDKN2C | 1416868_at 1439164_at | 21808 | -3.419 | -0.1009 | Yes | ||
| 138 | EGR2 | 1427682_a_at 1427683_at | 21875 | -4.015 | -0.0523 | Yes | ||
| 139 | RBM8A | 1418119_at 1418120_at | 21892 | -4.271 | 0.0020 | Yes |