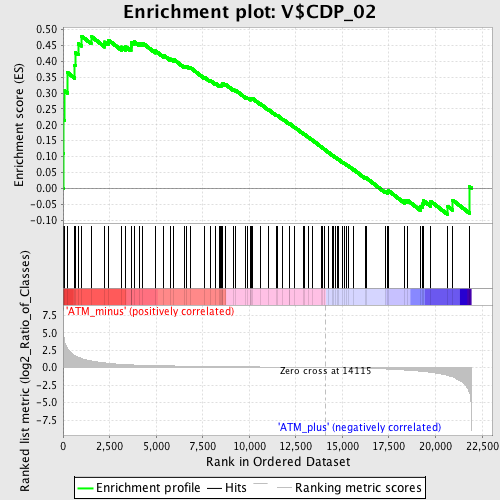
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$CDP_02 |
| Enrichment Score (ES) | 0.47921258 |
| Normalized Enrichment Score (NES) | 1.4298149 |
| Nominal p-value | 0.016427105 |
| FDR q-value | 0.38702688 |
| FWER p-Value | 0.98 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | WDR12 | 1417270_at 1448646_at 1459762_x_at | 31 | 4.677 | 0.1102 | Yes | ||
| 2 | MBNL1 | 1416904_at 1439686_at 1440315_at 1445106_at 1457924_at | 43 | 4.392 | 0.2145 | Yes | ||
| 3 | CDCA7 | 1428069_at 1445681_at | 66 | 3.948 | 0.3077 | Yes | ||
| 4 | CNIH | 1423727_at | 244 | 2.701 | 0.3641 | Yes | ||
| 5 | DOCK4 | 1431114_at 1436405_at 1441462_at 1441469_at 1446283_at 1457146_at 1459279_at | 626 | 1.741 | 0.3882 | Yes | ||
| 6 | JARID2 | 1422698_s_at 1446082_at 1450710_at 1456661_at | 656 | 1.698 | 0.4274 | Yes | ||
| 7 | DACH1 | 1420694_a_at 1420695_at 1433743_at 1447174_at 1459381_at | 818 | 1.490 | 0.4556 | Yes | ||
| 8 | CS | 1422577_at 1422578_at 1450667_a_at | 987 | 1.312 | 0.4792 | Yes | ||
| 9 | OGG1 | 1430078_a_at 1448815_at | 1505 | 0.964 | 0.4786 | No | ||
| 10 | SLC6A9 | 1417636_at 1431812_a_at | 2244 | 0.670 | 0.4608 | No | ||
| 11 | HNRPA2B1 | 1420365_a_at 1433829_a_at 1433830_at 1434047_x_at | 2438 | 0.614 | 0.4666 | No | ||
| 12 | IGSF4 | 1417376_a_at 1417377_at 1417378_at 1431611_a_at | 3151 | 0.461 | 0.4450 | No | ||
| 13 | KIF1B | 1423994_at 1423995_at 1425270_at 1451200_at 1451642_at 1451762_a_at 1455182_at 1458426_at | 3369 | 0.429 | 0.4453 | No | ||
| 14 | NDST4 | 1421318_at 1431452_at 1450195_at | 3648 | 0.392 | 0.4419 | No | ||
| 15 | FOXD3 | 1422210_at | 3652 | 0.391 | 0.4511 | No | ||
| 16 | POU3F4 | 1422164_at 1422165_at | 3665 | 0.390 | 0.4599 | No | ||
| 17 | SLC5A10 | 1440834_at | 3812 | 0.374 | 0.4621 | No | ||
| 18 | GNAO1 | 1421152_a_at 1448031_at 1460383_at | 4115 | 0.343 | 0.4565 | No | ||
| 19 | MAB21L2 | 1418934_at | 4272 | 0.332 | 0.4573 | No | ||
| 20 | TOB1 | 1423176_at 1440844_at | 4958 | 0.284 | 0.4327 | No | ||
| 21 | SREBF2 | 1426744_at 1452174_at | 5393 | 0.263 | 0.4192 | No | ||
| 22 | SEMA5B | 1422146_at 1435963_at | 5782 | 0.245 | 0.4073 | No | ||
| 23 | PTMA | 1423455_at | 5941 | 0.238 | 0.4057 | No | ||
| 24 | HOXA11 | 1420414_at | 6530 | 0.215 | 0.3839 | No | ||
| 25 | PDZRN4 | 1456512_at | 6634 | 0.210 | 0.3842 | No | ||
| 26 | HOXA7 | 1449499_at | 6814 | 0.203 | 0.3809 | No | ||
| 27 | ANKRD2 | 1419621_at | 7595 | 0.176 | 0.3494 | No | ||
| 28 | A2BP1 | 1418314_a_at 1438217_at 1447735_x_at 1455358_at 1456921_at | 7915 | 0.167 | 0.3388 | No | ||
| 29 | PHACTR3 | 1429651_at | 8204 | 0.159 | 0.3294 | No | ||
| 30 | CACNA2D3 | 1419225_at | 8417 | 0.152 | 0.3233 | No | ||
| 31 | GRIA3 | 1420563_at 1434728_at | 8463 | 0.151 | 0.3249 | No | ||
| 32 | SOX9 | 1424950_at 1433889_at 1451538_at | 8531 | 0.148 | 0.3253 | No | ||
| 33 | SEMA6A | 1421414_a_at 1425903_at 1436458_at | 8553 | 0.148 | 0.3279 | No | ||
| 34 | ERG | 1425370_a_at 1440244_at | 8564 | 0.148 | 0.3310 | No | ||
| 35 | MEF2C | 1421027_a_at 1421028_a_at 1424852_at 1439946_at 1445420_at 1446484_at 1451506_at 1451507_at 1458551_at | 8699 | 0.144 | 0.3283 | No | ||
| 36 | LRCH2 | 1436877_at 1441273_at | 9159 | 0.131 | 0.3104 | No | ||
| 37 | ETV4 | 1423232_at 1443381_at | 9279 | 0.128 | 0.3080 | No | ||
| 38 | SIX1 | 1427277_at | 9821 | 0.113 | 0.2859 | No | ||
| 39 | LSAMP | 1443754_x_at 1455636_at 1460092_at | 9926 | 0.110 | 0.2838 | No | ||
| 40 | TCF4 | 1416723_at 1416724_x_at 1416725_at 1424089_a_at 1434148_at 1434149_at 1439336_at 1440106_at 1446386_at 1458201_at | 10057 | 0.107 | 0.2804 | No | ||
| 41 | PTF1A | 1419424_at | 10081 | 0.106 | 0.2819 | No | ||
| 42 | PPM1L | 1435699_at 1435787_at 1438012_at | 10095 | 0.106 | 0.2838 | No | ||
| 43 | SYT1 | 1421990_at 1431191_a_at 1433884_at 1438282_at 1442557_at 1457497_at | 10193 | 0.103 | 0.2818 | No | ||
| 44 | SALL1 | 1450489_at | 10593 | 0.093 | 0.2658 | No | ||
| 45 | HCN1 | 1450193_at | 11048 | 0.082 | 0.2470 | No | ||
| 46 | EPHA3 | 1425574_at 1425575_at 1426057_a_at 1443273_at | 11462 | 0.071 | 0.2298 | No | ||
| 47 | GC | 1426547_at | 11509 | 0.070 | 0.2294 | No | ||
| 48 | PITX2 | 1424797_a_at 1450482_a_at | 11807 | 0.063 | 0.2173 | No | ||
| 49 | GPX1 | 1460671_at | 12158 | 0.054 | 0.2025 | No | ||
| 50 | PTGS2 | 1417262_at 1417263_at | 12170 | 0.054 | 0.2033 | No | ||
| 51 | CDH9 | 1427618_at | 12442 | 0.047 | 0.1921 | No | ||
| 52 | HOXA2 | 1419602_at | 12919 | 0.035 | 0.1711 | No | ||
| 53 | SLITRK2 | 1441483_at | 12960 | 0.033 | 0.1701 | No | ||
| 54 | SRPK2 | 1417134_at 1417135_at 1417136_s_at 1431372_at 1435746_at 1440080_at 1442241_at 1442980_at 1447755_at 1448603_at | 13199 | 0.027 | 0.1598 | No | ||
| 55 | LPHN2 | 1434111_at 1434112_at 1444906_at | 13407 | 0.021 | 0.1509 | No | ||
| 56 | NRXN3 | 1419825_at 1431153_at 1432931_at 1433788_at 1438193_at 1439629_at 1442423_at 1444700_at 1456137_at 1460101_at | 13873 | 0.007 | 0.1298 | No | ||
| 57 | MYH10 | 1441057_at 1452740_at | 13934 | 0.005 | 0.1272 | No | ||
| 58 | LIF | 1421206_at 1421207_at 1450160_at | 14044 | 0.002 | 0.1222 | No | ||
| 59 | BHLHB5 | 1418271_at | 14248 | -0.005 | 0.1130 | No | ||
| 60 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 14473 | -0.013 | 0.1031 | No | ||
| 61 | IL1RAPL1 | 1445821_at 1458964_at | 14520 | -0.015 | 0.1013 | No | ||
| 62 | TBX3 | 1437479_x_at 1439567_at 1444594_at 1445757_at 1448029_at | 14626 | -0.019 | 0.0970 | No | ||
| 63 | HOXB8 | 1452493_s_at | 14744 | -0.023 | 0.0922 | No | ||
| 64 | FGF9 | 1420795_at 1438718_at 1445258_at | 14774 | -0.024 | 0.0914 | No | ||
| 65 | DGKB | 1440901_at 1442860_at 1443631_at 1455361_at | 15009 | -0.033 | 0.0815 | No | ||
| 66 | LCP1 | 1415983_at 1448160_at | 15107 | -0.038 | 0.0780 | No | ||
| 67 | GBX2 | 1420337_at | 15242 | -0.043 | 0.0729 | No | ||
| 68 | GPM6A | 1426442_at 1456741_s_at 1459303_at | 15313 | -0.045 | 0.0708 | No | ||
| 69 | OTX1 | 1437601_at | 15619 | -0.060 | 0.0582 | No | ||
| 70 | SP8 | 1440519_at | 16230 | -0.094 | 0.0326 | No | ||
| 71 | PKD2L2 | 1419422_at 1449469_at | 16282 | -0.098 | 0.0326 | No | ||
| 72 | HNF4G | 1422182_at 1450518_at 1460127_at | 17300 | -0.185 | -0.0095 | No | ||
| 73 | STAG2 | 1421849_at 1450396_at | 17433 | -0.202 | -0.0108 | No | ||
| 74 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 17459 | -0.205 | -0.0070 | No | ||
| 75 | GNAQ | 1428938_at 1428939_s_at 1428940_at 1429559_at 1446688_at 1447593_x_at 1450115_at 1455729_at 1458159_at | 18345 | -0.332 | -0.0396 | No | ||
| 76 | RPA3 | 1448938_at | 18476 | -0.353 | -0.0371 | No | ||
| 77 | GTF2IRD1 | 1451899_a_at 1460364_at | 19195 | -0.510 | -0.0578 | No | ||
| 78 | CPEB4 | 1420617_at 1420618_at 1449931_at | 19289 | -0.532 | -0.0493 | No | ||
| 79 | DUSP10 | 1417163_at 1417164_at | 19355 | -0.548 | -0.0392 | No | ||
| 80 | SLC1A3 | 1426340_at 1426341_at 1439072_at 1440491_at 1443749_x_at 1452031_at | 19737 | -0.667 | -0.0408 | No | ||
| 81 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 20652 | -1.096 | -0.0564 | No | ||
| 82 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 20924 | -1.313 | -0.0375 | No | ||
| 83 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21827 | -3.509 | 0.0050 | No |

