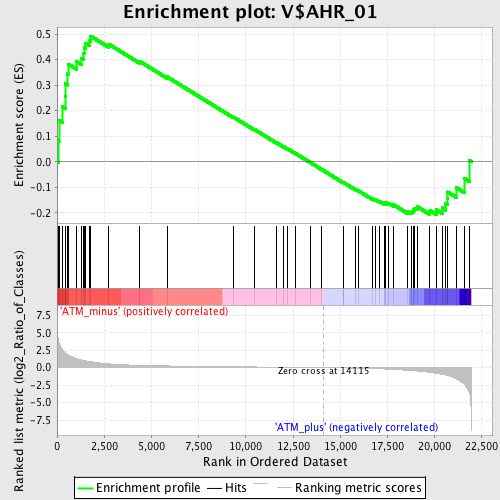
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$AHR_01 |
| Enrichment Score (ES) | 0.49199754 |
| Normalized Enrichment Score (NES) | 1.3521155 |
| Nominal p-value | 0.07630522 |
| FDR q-value | 0.42727897 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 82 | 3.710 | 0.0818 | Yes | ||
| 2 | DPYSL2 | 1433770_at 1450502_at 1458188_at 1458632_at | 101 | 3.510 | 0.1620 | Yes | ||
| 3 | ZFX | 1419565_a_at 1425972_a_at 1428046_a_at 1449512_a_at | 270 | 2.617 | 0.2147 | Yes | ||
| 4 | SUPT16H | 1419741_at 1449578_at 1456449_at | 425 | 2.144 | 0.2571 | Yes | ||
| 5 | LMO4 | 1420981_a_at 1439651_at | 439 | 2.111 | 0.3052 | Yes | ||
| 6 | HIST1H3D | 1427863_at 1453573_at | 545 | 1.880 | 0.3438 | Yes | ||
| 7 | CCND1 | 1417419_at 1417420_at 1448698_at | 613 | 1.756 | 0.3812 | Yes | ||
| 8 | RTN4 | 1421116_a_at 1435284_at 1437224_at 1439650_at 1442760_x_at 1452649_at | 1024 | 1.285 | 0.3921 | Yes | ||
| 9 | GARS | 1423784_at | 1274 | 1.098 | 0.4061 | Yes | ||
| 10 | MORF4L2 | 1415778_at 1439413_x_at | 1381 | 1.024 | 0.4249 | Yes | ||
| 11 | RUNX3 | 1421467_at 1440275_at | 1431 | 1.003 | 0.4458 | Yes | ||
| 12 | KPNB1 | 1416925_at 1434357_a_at 1448526_at 1451967_x_at | 1504 | 0.964 | 0.4647 | Yes | ||
| 13 | H1F0 | 1423702_at 1450522_a_at | 1726 | 0.865 | 0.4746 | Yes | ||
| 14 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 1772 | 0.844 | 0.4920 | Yes | ||
| 15 | PRX | 1423292_a_at | 2730 | 0.542 | 0.4608 | No | ||
| 16 | NR3C1 | 1421866_at 1421867_at 1453860_s_at 1457635_s_at 1460303_at | 4355 | 0.324 | 0.3941 | No | ||
| 17 | RPL10A | 1431177_a_at | 5825 | 0.244 | 0.3326 | No | ||
| 18 | SLC22A8 | 1416966_at 1435418_at | 9340 | 0.126 | 0.1749 | No | ||
| 19 | GRIK3 | 1427709_at | 10430 | 0.098 | 0.1274 | No | ||
| 20 | ELAVL3 | 1452524_a_at | 11633 | 0.067 | 0.0740 | No | ||
| 21 | KCNA6 | 1421568_at 1441049_at 1456954_at | 11996 | 0.058 | 0.0588 | No | ||
| 22 | NEUROG2 | 1422839_at | 12212 | 0.053 | 0.0502 | No | ||
| 23 | NTRK3 | 1422329_a_at 1425070_at 1425071_s_at 1426003_at 1433825_at 1443970_at 1455917_at | 12608 | 0.043 | 0.0332 | No | ||
| 24 | HMG20B | 1417637_a_at | 13413 | 0.021 | -0.0031 | No | ||
| 25 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 13998 | 0.004 | -0.0297 | No | ||
| 26 | SENP3 | 1448866_at | 15145 | -0.039 | -0.0811 | No | ||
| 27 | CHRM1 | 1439611_at 1450833_at | 15171 | -0.040 | -0.0814 | No | ||
| 28 | VAMP3 | 1421102_a_at 1433693_x_at 1433916_at 1437708_x_at 1456245_x_at 1457391_at | 15814 | -0.070 | -0.1091 | No | ||
| 29 | CYP1A1 | 1422217_a_at | 15974 | -0.079 | -0.1145 | No | ||
| 30 | NRF1 | 1420978_at 1424787_a_at 1434627_at 1439545_at 1445914_at | 16714 | -0.129 | -0.1453 | No | ||
| 31 | TGIF2 | 1431115_at 1434400_at | 16844 | -0.139 | -0.1479 | No | ||
| 32 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 17100 | -0.163 | -0.1558 | No | ||
| 33 | PAK6 | 1445475_at 1455200_at | 17358 | -0.192 | -0.1631 | No | ||
| 34 | EED | 1448653_at | 17402 | -0.198 | -0.1606 | No | ||
| 35 | POLD4 | 1427885_at | 17568 | -0.222 | -0.1630 | No | ||
| 36 | FBXW11 | 1425461_at 1425462_at 1437468_x_at 1438336_at 1443010_at 1447294_at 1457485_at | 17822 | -0.255 | -0.1687 | No | ||
| 37 | MNT | 1418192_at | 18560 | -0.368 | -0.1938 | No | ||
| 38 | PCBP2 | 1421743_a_at 1435881_at | 18795 | -0.417 | -0.1949 | No | ||
| 39 | PICALM | 1441328_at 1443218_at 1446968_at 1451316_a_at 1455773_at | 18881 | -0.435 | -0.1887 | No | ||
| 40 | PSCD2 | 1450103_a_at | 18944 | -0.450 | -0.1812 | No | ||
| 41 | NCAM1 | 1421966_at 1425126_at 1426864_a_at 1426865_a_at 1439556_at 1440067_at 1441995_at 1442680_at 1443018_at 1450437_a_at 1450438_at 1454140_at | 19097 | -0.487 | -0.1769 | No | ||
| 42 | POLG | 1423272_at 1423273_at 1437564_at | 19747 | -0.670 | -0.1911 | No | ||
| 43 | PANK3 | 1426259_at | 20078 | -0.792 | -0.1879 | No | ||
| 44 | ANGPTL2 | 1421002_at 1431848_at | 20400 | -0.941 | -0.1809 | No | ||
| 45 | TRIM39 | 1418851_at | 20587 | -1.049 | -0.1652 | No | ||
| 46 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20657 | -1.100 | -0.1430 | No | ||
| 47 | MKL1 | 1434900_at | 20686 | -1.116 | -0.1185 | No | ||
| 48 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 21159 | -1.642 | -0.1022 | No | ||
| 49 | CNOT10 | 1433882_at 1453654_at 1459756_at 1459757_x_at | 21591 | -2.419 | -0.0661 | No | ||
| 50 | EGR1 | 1417065_at | 21834 | -3.545 | 0.0047 | No |

