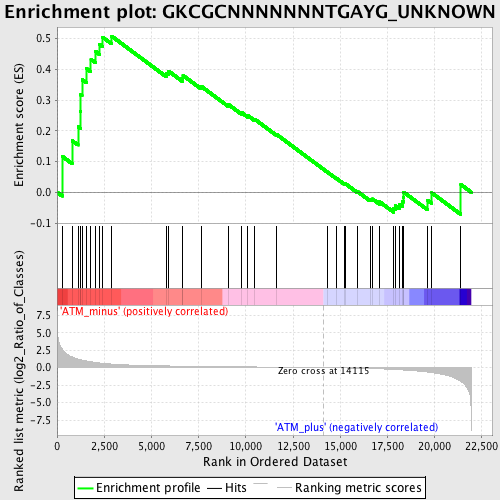
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | GKCGCNNNNNNNTGAYG_UNKNOWN |
| Enrichment Score (ES) | 0.5080695 |
| Normalized Enrichment Score (NES) | 1.3444822 |
| Nominal p-value | 0.08 |
| FDR q-value | 0.34427005 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RNF141 | 1418466_at 1427120_at 1433655_at 1449086_at 1449087_at | 269 | 2.622 | 0.1170 | Yes | ||
| 2 | COQ7 | 1416665_at | 796 | 1.523 | 0.1680 | Yes | ||
| 3 | PHF5A | 1424170_at | 1109 | 1.206 | 0.2132 | Yes | ||
| 4 | PHF7 | 1420260_at 1428659_at 1432386_a_at 1440828_x_at 1449800_x_at | 1231 | 1.125 | 0.2631 | Yes | ||
| 5 | PSME3 | 1418078_at 1418079_at 1438509_at | 1254 | 1.110 | 0.3169 | Yes | ||
| 6 | SENP2 | 1425465_a_at 1425466_at 1451717_s_at 1455327_at | 1321 | 1.073 | 0.3668 | Yes | ||
| 7 | ALS2CR2 | 1433544_at 1442452_at | 1559 | 0.939 | 0.4022 | Yes | ||
| 8 | GPAM | 1419499_at 1425834_a_at | 1793 | 0.833 | 0.4327 | Yes | ||
| 9 | UXT | 1418986_a_at | 2030 | 0.737 | 0.4582 | Yes | ||
| 10 | NUP155 | 1418727_at 1449200_at | 2256 | 0.666 | 0.4808 | Yes | ||
| 11 | PWP1 | 1417873_at 1454142_a_at | 2402 | 0.623 | 0.5048 | Yes | ||
| 12 | UBE4A | 1425278_at 1425384_a_at 1437747_at 1459708_at | 2884 | 0.511 | 0.5081 | Yes | ||
| 13 | LCMT1 | 1417044_at | 5790 | 0.245 | 0.3875 | No | ||
| 14 | BRMS1 | 1417897_at 1426129_at | 5908 | 0.240 | 0.3940 | No | ||
| 15 | MFN2 | 1441424_at 1448131_at 1451900_at | 6624 | 0.211 | 0.3717 | No | ||
| 16 | NUPL2 | 1424889_at 1456773_at | 6666 | 0.209 | 0.3802 | No | ||
| 17 | DHRS1 | 1415677_at | 7625 | 0.175 | 0.3450 | No | ||
| 18 | RCE1 | 1418779_at | 9080 | 0.133 | 0.2852 | No | ||
| 19 | DNAJB11 | 1423151_at | 9758 | 0.115 | 0.2600 | No | ||
| 20 | RNF7 | 1426414_a_at 1456600_a_at | 10098 | 0.106 | 0.2497 | No | ||
| 21 | NEK1 | 1434267_at 1438592_at 1453612_at 1458211_at | 10452 | 0.097 | 0.2383 | No | ||
| 22 | MRPL49 | 1423218_a_at 1423219_a_at | 11631 | 0.067 | 0.1878 | No | ||
| 23 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 14336 | -0.008 | 0.0647 | No | ||
| 24 | BAP1 | 1426354_at 1452039_a_at | 14809 | -0.026 | 0.0444 | No | ||
| 25 | NUP133 | 1423787_at 1423788_at 1436816_at 1441235_at 1451111_at 1457542_at | 15233 | -0.043 | 0.0272 | No | ||
| 26 | CNOT7 | 1423641_s_at 1430519_a_at 1460665_a_at | 15268 | -0.044 | 0.0278 | No | ||
| 27 | FAU | 1417452_a_at | 15884 | -0.074 | 0.0034 | No | ||
| 28 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 16578 | -0.118 | -0.0224 | No | ||
| 29 | RNPS1 | 1437027_x_at 1437359_at 1437851_x_at 1438267_x_at 1448324_at | 16703 | -0.128 | -0.0218 | No | ||
| 30 | QTRTD1 | 1421320_a_at 1451199_at | 17074 | -0.161 | -0.0307 | No | ||
| 31 | ORMDL3 | 1419450_at | 17817 | -0.254 | -0.0521 | No | ||
| 32 | RANBP2 | 1422621_at 1440104_at 1445883_at 1450690_at | 17929 | -0.269 | -0.0439 | No | ||
| 33 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18142 | -0.301 | -0.0387 | No | ||
| 34 | ANAPC10 | 1429375_at 1429376_s_at 1444033_at 1458390_at | 18296 | -0.324 | -0.0297 | No | ||
| 35 | BCL2L2 | 1423572_at 1430453_a_at 1430454_x_at 1451029_at | 18343 | -0.332 | -0.0154 | No | ||
| 36 | ABCE1 | 1416014_at 1416015_s_at 1442071_at | 18354 | -0.334 | 0.0006 | No | ||
| 37 | ACO2 | 1436934_s_at 1442102_at 1451002_at | 19611 | -0.623 | -0.0261 | No | ||
| 38 | NUP153 | 1441689_at 1441733_s_at 1452176_at | 19846 | -0.705 | -0.0020 | No | ||
| 39 | GTF3C2 | 1428448_a_at 1428449_at 1440307_at 1452780_at 1452781_a_at | 21386 | -1.975 | 0.0251 | No |

