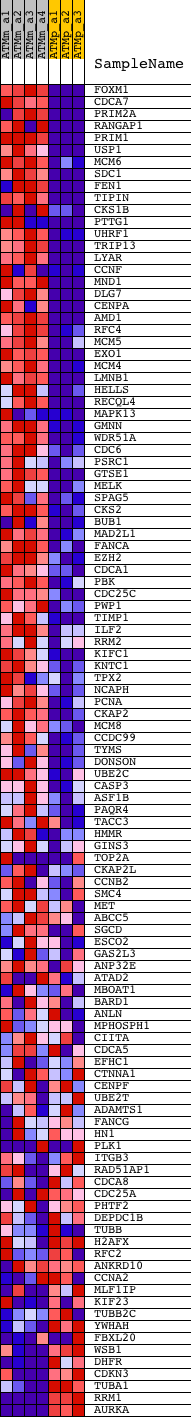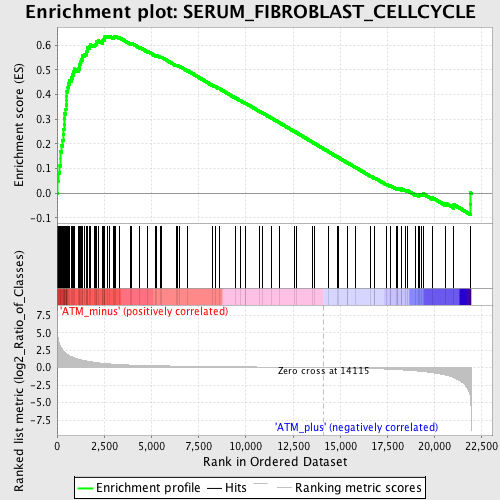
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | SERUM_FIBROBLAST_CELLCYCLE |
| Enrichment Score (ES) | 0.63604987 |
| Normalized Enrichment Score (NES) | 1.9693117 |
| Nominal p-value | 0.0 |
| FDR q-value | 8.500329E-4 |
| FWER p-Value | 0.0050 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FOXM1 | 1417748_x_at 1437138_at 1448833_at 1448834_at | 18 | 5.205 | 0.0492 | Yes | ||
| 2 | CDCA7 | 1428069_at 1445681_at | 66 | 3.948 | 0.0850 | Yes | ||
| 3 | PRIM2A | 1418035_a_at 1418036_at 1445474_at | 148 | 3.170 | 0.1117 | Yes | ||
| 4 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 184 | 3.018 | 0.1391 | Yes | ||
| 5 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 185 | 3.013 | 0.1681 | Yes | ||
| 6 | USP1 | 1423674_at 1423675_at 1451080_at | 229 | 2.819 | 0.1932 | Yes | ||
| 7 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 293 | 2.536 | 0.2147 | Yes | ||
| 8 | SDC1 | 1415943_at 1415944_at 1437279_x_at 1448158_at | 327 | 2.435 | 0.2366 | Yes | ||
| 9 | FEN1 | 1421731_a_at 1436454_x_at | 341 | 2.391 | 0.2590 | Yes | ||
| 10 | TIPIN | 1426612_at | 370 | 2.311 | 0.2799 | Yes | ||
| 11 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 383 | 2.275 | 0.3012 | Yes | ||
| 12 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 401 | 2.214 | 0.3217 | Yes | ||
| 13 | UHRF1 | 1415810_at 1415811_at 1439227_at | 464 | 2.046 | 0.3385 | Yes | ||
| 14 | TRIP13 | 1429294_at 1429295_s_at 1446760_at | 475 | 2.030 | 0.3576 | Yes | ||
| 15 | LYAR | 1417511_at | 507 | 1.958 | 0.3750 | Yes | ||
| 16 | CCNF | 1422513_at 1443807_x_at | 511 | 1.951 | 0.3936 | Yes | ||
| 17 | MND1 | 1452606_at | 516 | 1.946 | 0.4121 | Yes | ||
| 18 | DLG7 | 1438811_at 1455730_at | 572 | 1.823 | 0.4271 | Yes | ||
| 19 | CENPA | 1441864_x_at 1444416_at 1450842_a_at | 599 | 1.779 | 0.4430 | Yes | ||
| 20 | AMD1 | 1448484_at | 652 | 1.704 | 0.4570 | Yes | ||
| 21 | RFC4 | 1424321_at 1438161_s_at | 762 | 1.572 | 0.4671 | Yes | ||
| 22 | MCM5 | 1415945_at 1436808_x_at | 806 | 1.510 | 0.4796 | Yes | ||
| 23 | EXO1 | 1418026_at 1418027_at | 862 | 1.448 | 0.4910 | Yes | ||
| 24 | MCM4 | 1416214_at 1436708_x_at | 896 | 1.412 | 0.5031 | Yes | ||
| 25 | LMNB1 | 1423520_at 1423521_at | 1137 | 1.190 | 0.5035 | Yes | ||
| 26 | HELLS | 1417541_at 1430139_at 1453361_at | 1189 | 1.151 | 0.5123 | Yes | ||
| 27 | RECQL4 | 1422922_at | 1201 | 1.145 | 0.5228 | Yes | ||
| 28 | MAPK13 | 1448871_at | 1246 | 1.114 | 0.5315 | Yes | ||
| 29 | GMNN | 1417506_at | 1266 | 1.104 | 0.5412 | Yes | ||
| 30 | WDR51A | 1432202_a_at | 1333 | 1.068 | 0.5484 | Yes | ||
| 31 | CDC6 | 1417019_a_at | 1359 | 1.045 | 0.5573 | Yes | ||
| 32 | PSRC1 | 1417323_at 1425416_s_at 1451698_at | 1439 | 0.999 | 0.5633 | Yes | ||
| 33 | GTSE1 | 1416969_at | 1533 | 0.953 | 0.5682 | Yes | ||
| 34 | MELK | 1416558_at | 1576 | 0.933 | 0.5752 | Yes | ||
| 35 | SPAG5 | 1427498_a_at 1433892_at 1433893_s_at | 1606 | 0.918 | 0.5827 | Yes | ||
| 36 | CKS2 | 1417457_at 1417458_s_at | 1608 | 0.913 | 0.5915 | Yes | ||
| 37 | BUB1 | 1424046_at 1438571_at | 1704 | 0.876 | 0.5955 | Yes | ||
| 38 | MAD2L1 | 1422460_at | 1742 | 0.858 | 0.6021 | Yes | ||
| 39 | FANCA | 1418856_a_at 1452513_a_at | 1961 | 0.761 | 0.5994 | Yes | ||
| 40 | EZH2 | 1416544_at 1444263_at | 2029 | 0.737 | 0.6034 | Yes | ||
| 41 | CDCA1 | 1430811_a_at | 2081 | 0.720 | 0.6080 | Yes | ||
| 42 | PBK | 1448627_s_at | 2083 | 0.719 | 0.6149 | Yes | ||
| 43 | CDC25C | 1422252_a_at | 2169 | 0.693 | 0.6176 | Yes | ||
| 44 | PWP1 | 1417873_at 1454142_a_at | 2402 | 0.623 | 0.6130 | Yes | ||
| 45 | TIMP1 | 1460227_at | 2412 | 0.620 | 0.6185 | Yes | ||
| 46 | ILF2 | 1417948_s_at 1417949_at | 2443 | 0.611 | 0.6230 | Yes | ||
| 47 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 2521 | 0.591 | 0.6252 | Yes | ||
| 48 | KIFC1 | 1449877_s_at 1456136_at | 2530 | 0.589 | 0.6305 | Yes | ||
| 49 | KNTC1 | 1435575_at | 2533 | 0.588 | 0.6360 | Yes | ||
| 50 | TPX2 | 1428104_at 1428105_at 1447518_at 1447519_x_at | 2686 | 0.553 | 0.6344 | No | ||
| 51 | NCAPH | 1423920_at 1436707_x_at | 2785 | 0.529 | 0.6350 | No | ||
| 52 | PCNA | 1417947_at | 2980 | 0.492 | 0.6308 | No | ||
| 53 | CKAP2 | 1434748_at | 3015 | 0.485 | 0.6339 | No | ||
| 54 | MCM8 | 1429557_at | 3109 | 0.467 | 0.6342 | No | ||
| 55 | CCDC99 | 1424971_at | 3280 | 0.440 | 0.6306 | No | ||
| 56 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 3867 | 0.368 | 0.6073 | No | ||
| 57 | DONSON | 1426739_at | 3947 | 0.360 | 0.6071 | No | ||
| 58 | UBE2C | 1452954_at | 4374 | 0.323 | 0.5907 | No | ||
| 59 | CASP3 | 1426165_a_at 1430192_at 1449839_at | 4795 | 0.294 | 0.5743 | No | ||
| 60 | ASF1B | 1423714_at | 5224 | 0.271 | 0.5573 | No | ||
| 61 | PAQR4 | 1423101_at | 5282 | 0.269 | 0.5573 | No | ||
| 62 | TACC3 | 1417450_a_at 1436872_at 1455834_x_at | 5463 | 0.260 | 0.5515 | No | ||
| 63 | HMMR | 1425815_a_at 1427541_x_at 1429871_at 1450156_a_at 1450157_a_at | 5546 | 0.257 | 0.5502 | No | ||
| 64 | GINS3 | 1429149_at 1453050_at | 6314 | 0.223 | 0.5172 | No | ||
| 65 | TOP2A | 1427724_at 1442454_at 1454694_a_at | 6362 | 0.221 | 0.5172 | No | ||
| 66 | CKAP2L | 1435938_at 1453769_at | 6467 | 0.217 | 0.5145 | No | ||
| 67 | CCNB2 | 1450920_at | 6922 | 0.199 | 0.4956 | No | ||
| 68 | SMC4 | 1427275_at 1427276_at 1441677_at 1452197_at | 8221 | 0.158 | 0.4377 | No | ||
| 69 | MET | 1422990_at | 8369 | 0.153 | 0.4324 | No | ||
| 70 | ABCC5 | 1418042_a_at 1418043_at 1427565_a_at 1435683_a_at 1435684_at 1435685_x_at 1438056_x_at 1447384_at 1459782_x_at | 8582 | 0.147 | 0.4241 | No | ||
| 71 | SGCD | 1444822_at 1450805_at | 9426 | 0.123 | 0.3867 | No | ||
| 72 | ESCO2 | 1428304_at 1456311_x_at | 9710 | 0.116 | 0.3748 | No | ||
| 73 | GAS2L3 | 1437244_at 1453416_at 1455980_a_at | 9971 | 0.109 | 0.3640 | No | ||
| 74 | ANP32E | 1420592_a_at 1447680_at | 9974 | 0.109 | 0.3649 | No | ||
| 75 | ATAD2 | 1436174_at 1443146_at 1443229_at | 10715 | 0.091 | 0.3319 | No | ||
| 76 | MBOAT1 | 1435323_a_at 1441824_at | 10852 | 0.087 | 0.3265 | No | ||
| 77 | BARD1 | 1420594_at | 11351 | 0.074 | 0.3044 | No | ||
| 78 | ANLN | 1433543_at 1439648_at | 11764 | 0.064 | 0.2861 | No | ||
| 79 | MPHOSPH1 | 1419792_at 1439695_a_at 1440924_at 1449612_x_at | 12593 | 0.044 | 0.2486 | No | ||
| 80 | CIITA | 1421210_at 1421211_a_at 1447075_at | 12699 | 0.041 | 0.2442 | No | ||
| 81 | CDCA5 | 1416802_a_at 1448466_at | 13541 | 0.018 | 0.2058 | No | ||
| 82 | EFHC1 | 1453159_at | 13631 | 0.015 | 0.2019 | No | ||
| 83 | CTNNA1 | 1437275_at 1437807_x_at 1443662_at 1448149_at | 14357 | -0.009 | 0.1688 | No | ||
| 84 | CENPF | 1427161_at 1452334_at 1458447_at | 14868 | -0.028 | 0.1457 | No | ||
| 85 | UBE2T | 1422462_at 1442398_at | 14882 | -0.028 | 0.1454 | No | ||
| 86 | ADAMTS1 | 1450716_at | 15370 | -0.048 | 0.1235 | No | ||
| 87 | FANCG | 1420960_at | 15397 | -0.049 | 0.1228 | No | ||
| 88 | HN1 | 1416028_a_at 1438988_x_at 1448180_a_at | 15812 | -0.070 | 0.1045 | No | ||
| 89 | PLK1 | 1443408_at 1448191_at 1459616_at | 16609 | -0.121 | 0.0692 | No | ||
| 90 | ITGB3 | 1421511_at 1455257_at | 16828 | -0.138 | 0.0605 | No | ||
| 91 | RAD51AP1 | 1417938_at 1417939_at 1417940_s_at 1448899_s_at | 16833 | -0.139 | 0.0617 | No | ||
| 92 | CDCA8 | 1428480_at 1428481_s_at 1436847_s_at | 17438 | -0.202 | 0.0359 | No | ||
| 93 | CDC25A | 1417131_at 1417132_at 1445995_at | 17656 | -0.233 | 0.0282 | No | ||
| 94 | PHTF2 | 1437067_at 1437637_at 1444559_at | 17676 | -0.237 | 0.0296 | No | ||
| 95 | DEPDC1B | 1434789_at | 17976 | -0.274 | 0.0186 | No | ||
| 96 | TUBB | 1416256_a_at 1455719_at | 18028 | -0.282 | 0.0190 | No | ||
| 97 | H2AFX | 1416746_at | 18043 | -0.284 | 0.0210 | No | ||
| 98 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 18242 | -0.317 | 0.0150 | No | ||
| 99 | ANKRD10 | 1416065_a_at 1429304_at 1429305_at 1440170_at 1447832_x_at 1448199_at | 18246 | -0.318 | 0.0179 | No | ||
| 100 | CCNA2 | 1417910_at 1417911_at | 18439 | -0.346 | 0.0125 | No | ||
| 101 | MLF1IP | 1428518_at 1452816_at | 18568 | -0.371 | 0.0102 | No | ||
| 102 | KIF23 | 1450827_at 1453748_a_at 1455990_at | 18961 | -0.456 | -0.0034 | No | ||
| 103 | TUBB2C | 1423642_at 1438622_x_at 1439416_x_at 1456031_at 1456078_x_at 1456470_x_at | 19161 | -0.503 | -0.0077 | No | ||
| 104 | YWHAH | 1416004_at | 19192 | -0.509 | -0.0042 | No | ||
| 105 | FBXL20 | 1442464_at 1445575_at 1452826_s_at 1456378_s_at | 19319 | -0.540 | -0.0048 | No | ||
| 106 | WSB1 | 1425241_a_at | 19384 | -0.557 | -0.0024 | No | ||
| 107 | DHFR | 1419172_at 1430750_at | 19889 | -0.717 | -0.0186 | No | ||
| 108 | CDKN3 | 1415968_a_at 1415969_s_at 1430574_at | 20567 | -1.036 | -0.0396 | No | ||
| 109 | TUBA1 | 1418884_x_at | 21017 | -1.439 | -0.0464 | No | ||
| 110 | RRM1 | 1415878_at 1440073_at 1448127_at | 21883 | -4.186 | -0.0458 | No | ||
| 111 | AURKA | 1424511_at 1440129_at | 21907 | -5.010 | 0.0013 | No |