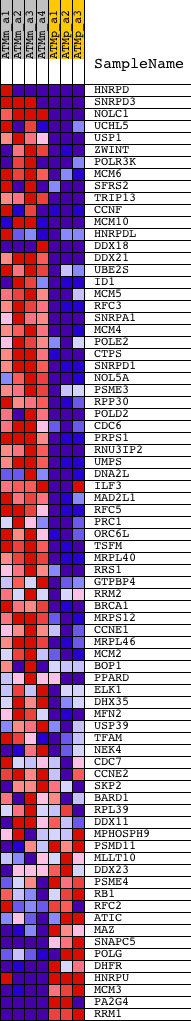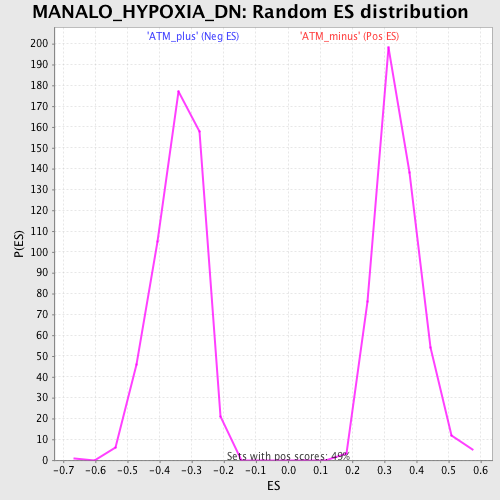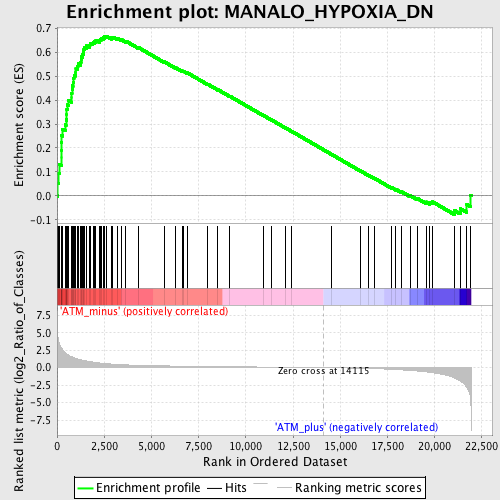
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | MANALO_HYPOXIA_DN |
| Enrichment Score (ES) | 0.667459 |
| Normalized Enrichment Score (NES) | 1.9446915 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.001156032 |
| FWER p-Value | 0.0090 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HNRPD | 1425142_a_at 1446083_at | 33 | 4.652 | 0.0515 | Yes | ||
| 2 | SNRPD3 | 1422884_at 1422885_at 1449629_s_at | 70 | 3.889 | 0.0941 | Yes | ||
| 3 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 110 | 3.462 | 0.1318 | Yes | ||
| 4 | UCHL5 | 1419452_at 1419453_at 1455086_at 1459011_at | 228 | 2.826 | 0.1586 | Yes | ||
| 5 | USP1 | 1423674_at 1423675_at 1451080_at | 229 | 2.819 | 0.1907 | Yes | ||
| 6 | ZWINT | 1423724_at 1427539_a_at 1427540_at 1429786_a_at 1429787_x_at 1444717_at 1455382_at | 236 | 2.731 | 0.2216 | Yes | ||
| 7 | POLR3K | 1422752_at 1422753_a_at 1439266_a_at 1440735_at 1450737_at 1456731_x_at | 248 | 2.697 | 0.2518 | Yes | ||
| 8 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 293 | 2.536 | 0.2787 | Yes | ||
| 9 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 428 | 2.140 | 0.2969 | Yes | ||
| 10 | TRIP13 | 1429294_at 1429295_s_at 1446760_at | 475 | 2.030 | 0.3179 | Yes | ||
| 11 | CCNF | 1422513_at 1443807_x_at | 511 | 1.951 | 0.3385 | Yes | ||
| 12 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 517 | 1.943 | 0.3604 | Yes | ||
| 13 | HNRPDL | 1420093_s_at 1420094_at 1424251_a_at 1424252_at 1428224_at 1428225_s_at 1449039_a_at 1456698_s_at | 554 | 1.864 | 0.3800 | Yes | ||
| 14 | DDX18 | 1416070_a_at 1416071_at 1456000_at | 593 | 1.790 | 0.3987 | Yes | ||
| 15 | DDX21 | 1448270_at 1448271_a_at | 747 | 1.586 | 0.4097 | Yes | ||
| 16 | UBE2S | 1416726_s_at 1430962_at | 749 | 1.585 | 0.4278 | Yes | ||
| 17 | ID1 | 1425895_a_at | 790 | 1.529 | 0.4433 | Yes | ||
| 18 | MCM5 | 1415945_at 1436808_x_at | 806 | 1.510 | 0.4599 | Yes | ||
| 19 | RFC3 | 1423700_at 1432538_a_at | 843 | 1.469 | 0.4749 | Yes | ||
| 20 | SNRPA1 | 1417351_a_at 1417352_s_at 1417353_x_at | 882 | 1.425 | 0.4894 | Yes | ||
| 21 | MCM4 | 1416214_at 1436708_x_at | 896 | 1.412 | 0.5049 | Yes | ||
| 22 | POLE2 | 1427094_at 1431440_at | 991 | 1.311 | 0.5156 | Yes | ||
| 23 | CTPS | 1416563_at | 998 | 1.305 | 0.5302 | Yes | ||
| 24 | SNRPD1 | 1416336_s_at | 1086 | 1.226 | 0.5401 | Yes | ||
| 25 | NOL5A | 1426533_at 1455035_s_at | 1108 | 1.209 | 0.5530 | Yes | ||
| 26 | PSME3 | 1418078_at 1418079_at 1438509_at | 1254 | 1.110 | 0.5590 | Yes | ||
| 27 | RPP30 | 1423373_at | 1268 | 1.102 | 0.5709 | Yes | ||
| 28 | POLD2 | 1448277_at | 1306 | 1.081 | 0.5816 | Yes | ||
| 29 | CDC6 | 1417019_a_at | 1359 | 1.045 | 0.5911 | Yes | ||
| 30 | PRPS1 | 1416052_at | 1386 | 1.021 | 0.6015 | Yes | ||
| 31 | RNU3IP2 | 1451293_at | 1417 | 1.008 | 0.6116 | Yes | ||
| 32 | UMPS | 1431652_at 1434859_at 1451445_at 1455832_a_at | 1449 | 0.991 | 0.6215 | Yes | ||
| 33 | DNA2L | 1438817_at 1452210_at 1457909_at | 1572 | 0.934 | 0.6266 | Yes | ||
| 34 | ILF3 | 1422546_at 1436802_at 1460669_at | 1711 | 0.873 | 0.6302 | Yes | ||
| 35 | MAD2L1 | 1422460_at | 1742 | 0.858 | 0.6386 | Yes | ||
| 36 | RFC5 | 1452917_at | 1900 | 0.786 | 0.6404 | Yes | ||
| 37 | PRC1 | 1423774_a_at 1423775_s_at | 1991 | 0.750 | 0.6448 | Yes | ||
| 38 | ORC6L | 1417037_at | 2054 | 0.727 | 0.6502 | Yes | ||
| 39 | TSFM | 1432170_at 1452698_at | 2253 | 0.667 | 0.6488 | Yes | ||
| 40 | MRPL40 | 1448849_at | 2285 | 0.656 | 0.6548 | Yes | ||
| 41 | RRS1 | 1416998_at 1456865_x_at | 2367 | 0.632 | 0.6583 | Yes | ||
| 42 | GTPBP4 | 1423142_a_at 1423143_at 1450873_at | 2440 | 0.613 | 0.6620 | Yes | ||
| 43 | RRM2 | 1416120_at 1434437_x_at 1448226_at | 2521 | 0.591 | 0.6651 | Yes | ||
| 44 | BRCA1 | 1424629_at 1424630_a_at 1427698_at 1443763_at 1451417_at | 2612 | 0.570 | 0.6675 | Yes | ||
| 45 | MRPS12 | 1448799_s_at | 2885 | 0.511 | 0.6608 | No | ||
| 46 | CCNE1 | 1416492_at 1441910_x_at | 2955 | 0.495 | 0.6633 | No | ||
| 47 | MRPL46 | 1449004_at | 3211 | 0.450 | 0.6568 | No | ||
| 48 | MCM2 | 1434079_s_at 1448777_at | 3383 | 0.426 | 0.6538 | No | ||
| 49 | BOP1 | 1423264_at 1430491_at | 3642 | 0.393 | 0.6465 | No | ||
| 50 | PPARD | 1425703_at 1439797_at | 4309 | 0.329 | 0.6198 | No | ||
| 51 | ELK1 | 1421896_at 1421897_at 1446390_at | 5676 | 0.250 | 0.5601 | No | ||
| 52 | DHX35 | 1452693_at | 6272 | 0.224 | 0.5355 | No | ||
| 53 | MFN2 | 1441424_at 1448131_at 1451900_at | 6624 | 0.211 | 0.5218 | No | ||
| 54 | USP39 | 1437007_x_at 1460209_at | 6714 | 0.207 | 0.5201 | No | ||
| 55 | TFAM | 1448224_at 1456215_at | 6902 | 0.200 | 0.5138 | No | ||
| 56 | NEK4 | 1427639_a_at 1449861_at | 7981 | 0.165 | 0.4664 | No | ||
| 57 | CDC7 | 1426002_a_at 1426021_a_at | 8519 | 0.149 | 0.4435 | No | ||
| 58 | CCNE2 | 1422535_at | 9139 | 0.132 | 0.4167 | No | ||
| 59 | SKP2 | 1418969_at 1425072_at 1436000_a_at 1437033_a_at 1449293_a_at 1460247_a_at | 10906 | 0.086 | 0.3369 | No | ||
| 60 | BARD1 | 1420594_at | 11351 | 0.074 | 0.3174 | No | ||
| 61 | RPL39 | 1423032_at 1450840_a_at | 12112 | 0.055 | 0.2833 | No | ||
| 62 | DDX11 | 1438447_at 1441062_at | 12396 | 0.049 | 0.2709 | No | ||
| 63 | MPHOSPH9 | 1419909_at 1419910_at 1431053_at 1438063_at 1440915_at 1449659_s_at | 14557 | -0.016 | 0.1722 | No | ||
| 64 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 16069 | -0.085 | 0.1041 | No | ||
| 65 | MLLT10 | 1419772_at 1420869_at 1420870_at 1438525_at 1442070_at 1443500_at | 16485 | -0.111 | 0.0864 | No | ||
| 66 | DDX23 | 1430050_at | 16810 | -0.137 | 0.0731 | No | ||
| 67 | PSME4 | 1426823_s_at 1426824_at 1440511_at 1445147_at 1446464_at 1452211_at | 17724 | -0.243 | 0.0341 | No | ||
| 68 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 17944 | -0.270 | 0.0272 | No | ||
| 69 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 18242 | -0.317 | 0.0172 | No | ||
| 70 | ATIC | 1428506_at 1452811_at | 18710 | -0.397 | 0.0003 | No | ||
| 71 | MAZ | 1427099_at | 19064 | -0.479 | -0.0104 | No | ||
| 72 | SNAPC5 | 1427999_at | 19551 | -0.607 | -0.0257 | No | ||
| 73 | POLG | 1423272_at 1423273_at 1437564_at | 19747 | -0.670 | -0.0270 | No | ||
| 74 | DHFR | 1419172_at 1430750_at | 19889 | -0.717 | -0.0252 | No | ||
| 75 | HNRPU | 1423050_s_at 1423051_at 1434390_at 1434391_at 1450849_at | 21041 | -1.468 | -0.0612 | No | ||
| 76 | MCM3 | 1420029_at 1426653_at | 21359 | -1.933 | -0.0537 | No | ||
| 77 | PA2G4 | 1420142_s_at 1423060_at 1435372_a_at 1450854_at 1460265_at | 21700 | -2.834 | -0.0369 | No | ||
| 78 | RRM1 | 1415878_at 1440073_at 1448127_at | 21883 | -4.186 | 0.0024 | No |