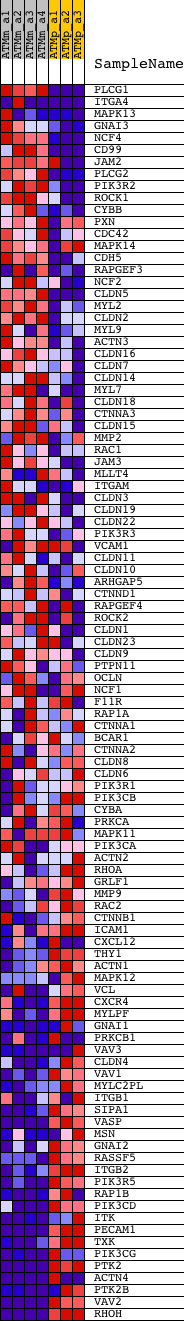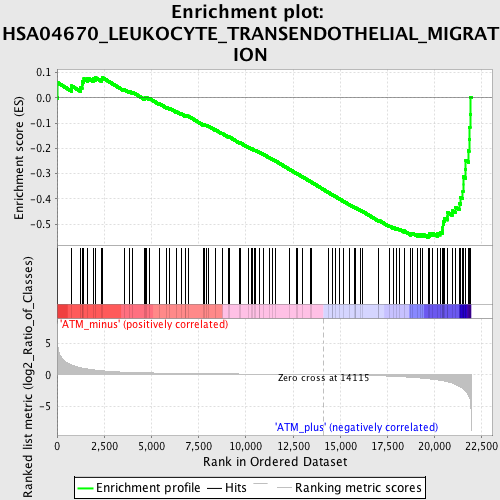
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | HSA04670_LEUKOCYTE_TRANSENDOTHELIAL_MIGRATION |
| Enrichment Score (ES) | -0.553051 |
| Normalized Enrichment Score (NES) | -1.6877235 |
| Nominal p-value | 0.0019157088 |
| FDR q-value | 0.16800222 |
| FWER p-Value | 0.952 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PLCG1 | 1435149_at 1450360_at | 37 | 4.528 | 0.0594 | No | ||
| 2 | ITGA4 | 1421194_at 1427615_at 1446350_at 1450155_at 1456498_at 1457376_at | 767 | 1.567 | 0.0471 | No | ||
| 3 | MAPK13 | 1448871_at | 1246 | 1.114 | 0.0402 | No | ||
| 4 | GNAI3 | 1428645_at 1437225_x_at | 1322 | 1.073 | 0.0513 | No | ||
| 5 | NCF4 | 1418465_at | 1328 | 1.069 | 0.0654 | No | ||
| 6 | CD99 | 1428850_x_at 1430514_a_at 1453556_x_at | 1420 | 1.006 | 0.0749 | No | ||
| 7 | JAM2 | 1419288_at 1431416_a_at 1431417_at 1436568_at 1449408_at | 1624 | 0.908 | 0.0778 | No | ||
| 8 | PLCG2 | 1426926_at | 1909 | 0.782 | 0.0753 | No | ||
| 9 | PIK3R2 | 1418463_at | 2025 | 0.739 | 0.0800 | No | ||
| 10 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 2376 | 0.630 | 0.0725 | No | ||
| 11 | CYBB | 1422978_at 1436778_at 1436779_at | 2400 | 0.623 | 0.0798 | No | ||
| 12 | PXN | 1424027_at 1426085_a_at 1456135_s_at | 3543 | 0.405 | 0.0330 | No | ||
| 13 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 3826 | 0.373 | 0.0251 | No | ||
| 14 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 4014 | 0.353 | 0.0213 | No | ||
| 15 | CDH5 | 1422047_at 1433956_at | 4626 | 0.304 | -0.0026 | No | ||
| 16 | RAPGEF3 | 1424470_a_at 1424471_at 1437012_x_at 1437946_x_at 1438590_at | 4656 | 0.302 | 0.0002 | No | ||
| 17 | NCF2 | 1448561_at | 4736 | 0.297 | 0.0006 | No | ||
| 18 | CLDN5 | 1417839_at | 4917 | 0.287 | -0.0038 | No | ||
| 19 | MYL2 | 1448394_at | 5422 | 0.262 | -0.0234 | No | ||
| 20 | CLDN2 | 1417231_at 1417232_at | 5818 | 0.244 | -0.0382 | No | ||
| 21 | MYL9 | 1452670_at | 5965 | 0.237 | -0.0417 | No | ||
| 22 | ACTN3 | 1418677_at | 6299 | 0.224 | -0.0539 | No | ||
| 23 | CLDN16 | 1420434_at | 6562 | 0.214 | -0.0630 | No | ||
| 24 | CLDN7 | 1448393_at | 6804 | 0.204 | -0.0713 | No | ||
| 25 | CLDN14 | 1420345_at | 6811 | 0.203 | -0.0688 | No | ||
| 26 | MYL7 | 1449071_at | 6940 | 0.199 | -0.0720 | No | ||
| 27 | CLDN18 | 1425445_a_at 1449428_at | 7775 | 0.171 | -0.1079 | No | ||
| 28 | CTNNA3 | 1441560_at | 7788 | 0.171 | -0.1062 | No | ||
| 29 | CLDN15 | 1418920_at | 7895 | 0.167 | -0.1088 | No | ||
| 30 | MMP2 | 1416136_at 1439364_a_at | 8034 | 0.163 | -0.1129 | No | ||
| 31 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 8362 | 0.154 | -0.1258 | No | ||
| 32 | JAM3 | 1423503_at 1423504_at 1444895_at | 8744 | 0.142 | -0.1413 | No | ||
| 33 | MLLT4 | 1431799_at 1436303_at | 9068 | 0.133 | -0.1543 | No | ||
| 34 | ITGAM | 1422046_at | 9122 | 0.132 | -0.1550 | No | ||
| 35 | CLDN3 | 1426332_a_at 1434651_a_at 1451701_x_at 1453258_at 1460569_x_at | 9665 | 0.117 | -0.1782 | No | ||
| 36 | CLDN19 | 1425727_at | 9691 | 0.117 | -0.1778 | No | ||
| 37 | CLDN22 | 1430237_at | 10135 | 0.105 | -0.1967 | No | ||
| 38 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 10269 | 0.101 | -0.2014 | No | ||
| 39 | VCAM1 | 1415989_at 1436003_at 1448162_at 1451314_a_at | 10324 | 0.100 | -0.2025 | No | ||
| 40 | CLDN11 | 1416003_at | 10454 | 0.097 | -0.2071 | No | ||
| 41 | CLDN10 | 1426147_s_at | 10495 | 0.096 | -0.2076 | No | ||
| 42 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 10717 | 0.091 | -0.2165 | No | ||
| 43 | CTNND1 | 1422450_at 1437448_s_at 1445830_at | 10744 | 0.090 | -0.2165 | No | ||
| 44 | RAPGEF4 | 1421622_a_at 1425518_at 1440274_at 1444034_at | 10945 | 0.085 | -0.2245 | No | ||
| 45 | ROCK2 | 1423592_at 1451041_at | 11226 | 0.078 | -0.2363 | No | ||
| 46 | CLDN1 | 1437932_a_at 1438850_at 1438851_x_at 1450014_at | 11399 | 0.073 | -0.2432 | No | ||
| 47 | CLDN23 | 1424409_at | 11568 | 0.069 | -0.2500 | No | ||
| 48 | CLDN9 | 1439427_at 1450524_at | 12314 | 0.051 | -0.2834 | No | ||
| 49 | PTPN11 | 1421196_at 1427699_a_at 1451225_at | 12683 | 0.041 | -0.2997 | No | ||
| 50 | OCLN | 1448873_at | 12736 | 0.040 | -0.3015 | No | ||
| 51 | NCF1 | 1425609_at 1451767_at 1456772_at | 12997 | 0.033 | -0.3130 | No | ||
| 52 | F11R | 1424595_at 1436374_x_at | 13437 | 0.021 | -0.3328 | No | ||
| 53 | RAP1A | 1424139_at | 13497 | 0.019 | -0.3353 | No | ||
| 54 | CTNNA1 | 1437275_at 1437807_x_at 1443662_at 1448149_at | 14357 | -0.009 | -0.3745 | No | ||
| 55 | BCAR1 | 1439388_s_at 1450622_at | 14577 | -0.017 | -0.3843 | No | ||
| 56 | CTNNA2 | 1442893_at 1447117_at 1448895_a_at | 14578 | -0.017 | -0.3841 | No | ||
| 57 | CLDN8 | 1449091_at | 14725 | -0.022 | -0.3905 | No | ||
| 58 | CLDN6 | 1417845_at | 14937 | -0.031 | -0.3997 | No | ||
| 59 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 15150 | -0.039 | -0.4089 | No | ||
| 60 | PIK3CB | 1445731_at 1453069_at | 15462 | -0.052 | -0.4224 | No | ||
| 61 | CYBA | 1454268_a_at | 15745 | -0.067 | -0.4345 | No | ||
| 62 | PRKCA | 1427562_a_at 1443069_at 1445028_at 1445395_at 1446598_at 1450945_at | 15821 | -0.071 | -0.4369 | No | ||
| 63 | MAPK11 | 1421925_at 1421926_at | 15824 | -0.071 | -0.4361 | No | ||
| 64 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 16083 | -0.086 | -0.4467 | No | ||
| 65 | ACTN2 | 1448327_at 1456968_at | 16197 | -0.093 | -0.4507 | No | ||
| 66 | RHOA | 1437628_s_at | 17032 | -0.156 | -0.4868 | No | ||
| 67 | GRLF1 | 1434009_at 1440675_at 1442181_at | 17046 | -0.157 | -0.4852 | No | ||
| 68 | MMP9 | 1416298_at 1448291_at | 17610 | -0.228 | -0.5079 | No | ||
| 69 | RAC2 | 1417620_at 1440208_at | 17826 | -0.255 | -0.5144 | No | ||
| 70 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 18001 | -0.278 | -0.5186 | No | ||
| 71 | ICAM1 | 1424067_at | 18113 | -0.296 | -0.5197 | No | ||
| 72 | CXCL12 | 1417574_at 1439084_at 1448823_at | 18377 | -0.337 | -0.5272 | No | ||
| 73 | THY1 | 1423135_at | 18740 | -0.402 | -0.5383 | No | ||
| 74 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 18819 | -0.422 | -0.5362 | No | ||
| 75 | MAPK12 | 1447823_x_at 1449283_a_at | 19094 | -0.486 | -0.5422 | No | ||
| 76 | VCL | 1416156_at 1416157_at 1445256_at | 19248 | -0.521 | -0.5422 | No | ||
| 77 | CXCR4 | 1448710_at | 19371 | -0.553 | -0.5403 | No | ||
| 78 | MYLPF | 1448371_at | 19650 | -0.638 | -0.5445 | Yes | ||
| 79 | GNAI1 | 1427510_at 1434440_at 1454959_s_at | 19723 | -0.662 | -0.5388 | Yes | ||
| 80 | PRKCB1 | 1423478_at 1438981_at 1443144_at 1459674_at 1460419_a_at | 19874 | -0.712 | -0.5361 | Yes | ||
| 81 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20163 | -0.833 | -0.5381 | Yes | ||
| 82 | CLDN4 | 1418283_at | 20280 | -0.882 | -0.5315 | Yes | ||
| 83 | VAV1 | 1422932_a_at | 20423 | -0.958 | -0.5250 | Yes | ||
| 84 | MYLC2PL | 1420805_at | 20432 | -0.962 | -0.5124 | Yes | ||
| 85 | ITGB1 | 1426918_at 1426919_at 1426920_x_at 1427771_x_at 1438119_at 1452545_a_at | 20438 | -0.966 | -0.4996 | Yes | ||
| 86 | SIPA1 | 1416206_at | 20481 | -0.983 | -0.4883 | Yes | ||
| 87 | VASP | 1451097_at | 20512 | -1.003 | -0.4762 | Yes | ||
| 88 | MSN | 1421814_at 1450379_at | 20683 | -1.114 | -0.4689 | Yes | ||
| 89 | GNAI2 | 1419449_a_at 1435652_a_at | 20687 | -1.116 | -0.4540 | Yes | ||
| 90 | RASSF5 | 1422637_at 1422638_s_at | 20921 | -1.311 | -0.4470 | Yes | ||
| 91 | ITGB2 | 1450678_at | 21110 | -1.563 | -0.4345 | Yes | ||
| 92 | PIK3R5 | 1434980_at | 21318 | -1.866 | -0.4189 | Yes | ||
| 93 | RAP1B | 1435518_at 1435519_at 1455349_at | 21373 | -1.951 | -0.3950 | Yes | ||
| 94 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 21492 | -2.164 | -0.3713 | Yes | ||
| 95 | ITK | 1417171_at 1430833_at 1452518_a_at 1456836_at 1457120_at | 21504 | -2.182 | -0.3423 | Yes | ||
| 96 | PECAM1 | 1421287_a_at 1447092_at | 21505 | -2.184 | -0.3129 | Yes | ||
| 97 | TXK | 1425785_a_at | 21628 | -2.579 | -0.2837 | Yes | ||
| 98 | PIK3CG | 1422707_at 1422708_at | 21647 | -2.651 | -0.2488 | Yes | ||
| 99 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 21796 | -3.373 | -0.2101 | Yes | ||
| 100 | ACTN4 | 1423449_a_at 1444258_at | 21819 | -3.472 | -0.1643 | Yes | ||
| 101 | PTK2B | 1434653_at 1442437_at 1442927_at | 21836 | -3.576 | -0.1168 | Yes | ||
| 102 | VAV2 | 1421272_at 1435065_x_at 1435244_at 1458758_at | 21871 | -3.956 | -0.0650 | Yes | ||
| 103 | RHOH | 1429319_at 1441385_at 1443015_at | 21909 | -5.037 | 0.0012 | Yes |