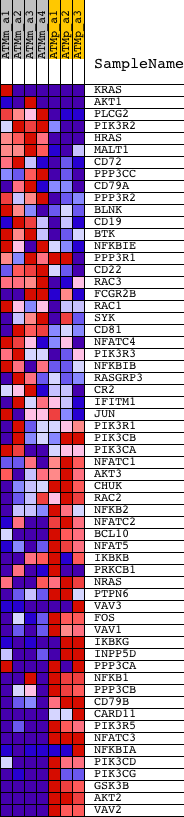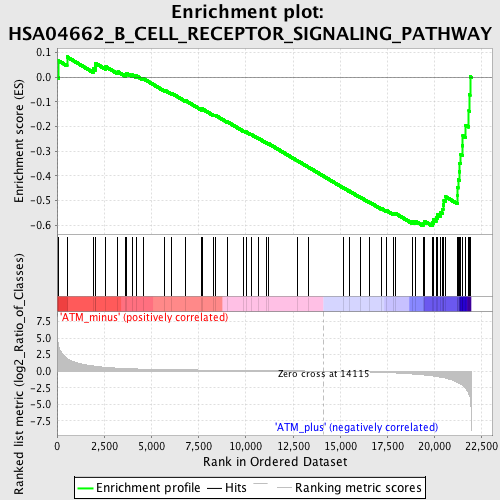
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | HSA04662_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.60287327 |
| Normalized Enrichment Score (NES) | -1.7323269 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.16718598 |
| FWER p-Value | 0.778 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KRAS | 1426228_at 1426229_s_at 1434000_at 1451979_at | 84 | 3.699 | 0.0664 | No | ||
| 2 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 525 | 1.916 | 0.0826 | No | ||
| 3 | PLCG2 | 1426926_at | 1909 | 0.782 | 0.0343 | No | ||
| 4 | PIK3R2 | 1418463_at | 2025 | 0.739 | 0.0430 | No | ||
| 5 | HRAS | 1422407_s_at 1424132_at | 2039 | 0.734 | 0.0564 | No | ||
| 6 | MALT1 | 1445068_at 1456126_at 1456429_at | 2583 | 0.576 | 0.0425 | No | ||
| 7 | CD72 | 1426112_a_at | 3223 | 0.448 | 0.0218 | No | ||
| 8 | PPP3CC | 1420743_a_at 1430025_at | 3647 | 0.392 | 0.0099 | No | ||
| 9 | CD79A | 1418830_at | 3670 | 0.389 | 0.0163 | No | ||
| 10 | PPP3R2 | 1437535_at | 3971 | 0.358 | 0.0094 | No | ||
| 11 | BLNK | 1451780_at | 4193 | 0.337 | 0.0057 | No | ||
| 12 | CD19 | 1450570_a_at | 4551 | 0.309 | -0.0048 | No | ||
| 13 | BTK | 1422755_at | 5693 | 0.249 | -0.0522 | No | ||
| 14 | NFKBIE | 1431843_a_at 1458299_s_at | 6075 | 0.232 | -0.0652 | No | ||
| 15 | PPP3R1 | 1421786_at 1433591_at 1450368_a_at | 6792 | 0.204 | -0.0941 | No | ||
| 16 | CD22 | 1419768_at 1419769_at 1445222_at | 7656 | 0.174 | -0.1302 | No | ||
| 17 | RAC3 | 1420554_a_at | 7674 | 0.174 | -0.1277 | No | ||
| 18 | FCGR2B | 1435476_a_at 1435477_s_at 1447622_at 1451941_a_at 1455332_x_at | 8308 | 0.155 | -0.1537 | No | ||
| 19 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 8362 | 0.154 | -0.1532 | No | ||
| 20 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 9007 | 0.135 | -0.1801 | No | ||
| 21 | CD81 | 1416330_at | 9895 | 0.111 | -0.2185 | No | ||
| 22 | NFATC4 | 1423380_s_at 1432821_at 1454369_a_at | 10010 | 0.108 | -0.2217 | No | ||
| 23 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 10269 | 0.101 | -0.2316 | No | ||
| 24 | NFKBIB | 1421266_s_at 1446718_at | 10653 | 0.092 | -0.2473 | No | ||
| 25 | RASGRP3 | 1438030_at 1438031_at | 11086 | 0.081 | -0.2655 | No | ||
| 26 | CR2 | 1425289_a_at | 11217 | 0.078 | -0.2700 | No | ||
| 27 | IFITM1 | 1424254_at | 12717 | 0.040 | -0.3377 | No | ||
| 28 | JUN | 1417409_at 1448694_at | 13314 | 0.024 | -0.3645 | No | ||
| 29 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 15150 | -0.039 | -0.4477 | No | ||
| 30 | PIK3CB | 1445731_at 1453069_at | 15462 | -0.052 | -0.4609 | No | ||
| 31 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 16083 | -0.086 | -0.4876 | No | ||
| 32 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 16559 | -0.117 | -0.5071 | No | ||
| 33 | AKT3 | 1422078_at 1435260_at 1435879_at 1443222_at 1460307_at | 17177 | -0.171 | -0.5321 | No | ||
| 34 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 17435 | -0.202 | -0.5400 | No | ||
| 35 | RAC2 | 1417620_at 1440208_at | 17826 | -0.255 | -0.5530 | No | ||
| 36 | NFKB2 | 1425902_a_at 1429128_x_at | 17945 | -0.270 | -0.5532 | No | ||
| 37 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 18829 | -0.424 | -0.5855 | No | ||
| 38 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 18974 | -0.459 | -0.5834 | No | ||
| 39 | NFAT5 | 1426029_a_at 1438999_a_at 1439328_at 1439805_at 1448966_a_at 1451921_a_at 1458213_at | 19385 | -0.557 | -0.5916 | Yes | ||
| 40 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 19447 | -0.577 | -0.5834 | Yes | ||
| 41 | PRKCB1 | 1423478_at 1438981_at 1443144_at 1459674_at 1460419_a_at | 19874 | -0.712 | -0.5894 | Yes | ||
| 42 | NRAS | 1422688_a_at 1454060_a_at | 19925 | -0.733 | -0.5777 | Yes | ||
| 43 | PTPN6 | 1456694_x_at 1460188_at | 20072 | -0.789 | -0.5694 | Yes | ||
| 44 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20163 | -0.833 | -0.5577 | Yes | ||
| 45 | FOS | 1423100_at | 20289 | -0.888 | -0.5466 | Yes | ||
| 46 | VAV1 | 1422932_a_at | 20423 | -0.958 | -0.5345 | Yes | ||
| 47 | IKBKG | 1421208_at 1421209_s_at 1435646_at 1435647_at 1450161_at 1454690_at | 20443 | -0.967 | -0.5170 | Yes | ||
| 48 | INPP5D | 1418110_a_at 1424195_a_at | 20487 | -0.986 | -0.5002 | Yes | ||
| 49 | PPP3CA | 1426401_at 1438478_a_at 1440051_at 1452056_s_at 1458495_at | 20582 | -1.046 | -0.4847 | Yes | ||
| 50 | NFKB1 | 1427705_a_at 1442949_at | 21207 | -1.718 | -0.4806 | Yes | ||
| 51 | PPP3CB | 1427468_at 1428473_at 1428474_at 1433835_at 1446149_at 1459378_at | 21215 | -1.729 | -0.4481 | Yes | ||
| 52 | CD79B | 1417640_at | 21236 | -1.761 | -0.4155 | Yes | ||
| 53 | CARD11 | 1430257_at 1435996_at | 21299 | -1.844 | -0.3834 | Yes | ||
| 54 | PIK3R5 | 1434980_at | 21318 | -1.866 | -0.3488 | Yes | ||
| 55 | NFATC3 | 1419976_s_at 1452497_a_at | 21381 | -1.965 | -0.3143 | Yes | ||
| 56 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 21477 | -2.135 | -0.2781 | Yes | ||
| 57 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 21492 | -2.164 | -0.2377 | Yes | ||
| 58 | PIK3CG | 1422707_at 1422708_at | 21647 | -2.651 | -0.1944 | Yes | ||
| 59 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21811 | -3.432 | -0.1367 | Yes | ||
| 60 | AKT2 | 1421324_a_at 1424480_s_at 1455703_at | 21832 | -3.540 | -0.0704 | Yes | ||
| 61 | VAV2 | 1421272_at 1435065_x_at 1435244_at 1458758_at | 21871 | -3.956 | 0.0030 | Yes |