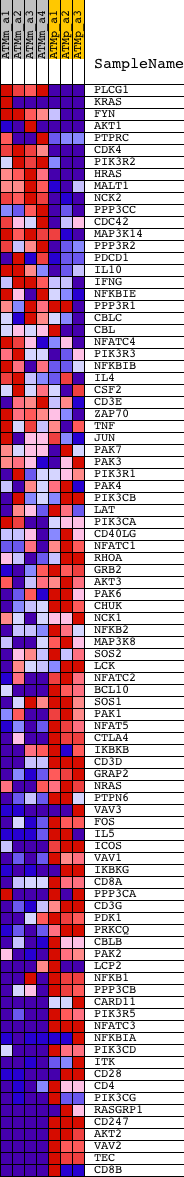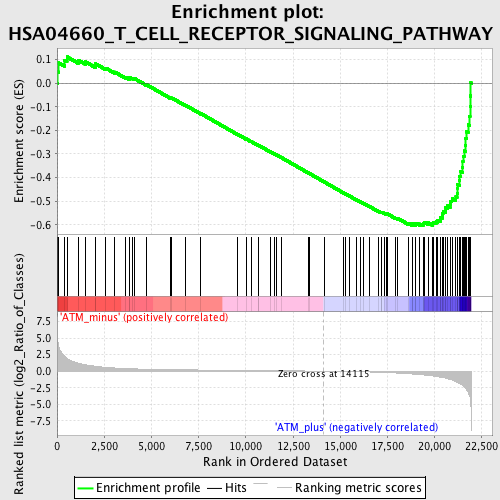
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | HSA04660_T_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.60468173 |
| Normalized Enrichment Score (NES) | -1.8092543 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07846209 |
| FWER p-Value | 0.335 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PLCG1 | 1435149_at 1450360_at | 37 | 4.528 | 0.0476 | No | ||
| 2 | KRAS | 1426228_at 1426229_s_at 1434000_at 1451979_at | 84 | 3.699 | 0.0858 | No | ||
| 3 | FYN | 1417558_at 1441647_at 1448765_at | 384 | 2.271 | 0.0968 | No | ||
| 4 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 525 | 1.916 | 0.1112 | No | ||
| 5 | PTPRC | 1422124_a_at 1440165_at | 1114 | 1.204 | 0.0974 | No | ||
| 6 | CDK4 | 1422441_x_at | 1506 | 0.963 | 0.0900 | No | ||
| 7 | PIK3R2 | 1418463_at | 2025 | 0.739 | 0.0743 | No | ||
| 8 | HRAS | 1422407_s_at 1424132_at | 2039 | 0.734 | 0.0817 | No | ||
| 9 | MALT1 | 1445068_at 1456126_at 1456429_at | 2583 | 0.576 | 0.0632 | No | ||
| 10 | NCK2 | 1416796_at 1416797_at 1458432_at | 3048 | 0.479 | 0.0471 | No | ||
| 11 | PPP3CC | 1420743_a_at 1430025_at | 3647 | 0.392 | 0.0240 | No | ||
| 12 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 3826 | 0.373 | 0.0199 | No | ||
| 13 | MAP3K14 | 1422999_at 1434364_at | 3846 | 0.371 | 0.0231 | No | ||
| 14 | PPP3R2 | 1437535_at | 3971 | 0.358 | 0.0213 | No | ||
| 15 | PDCD1 | 1449835_at | 4090 | 0.346 | 0.0197 | No | ||
| 16 | IL10 | 1450330_at | 4715 | 0.299 | -0.0056 | No | ||
| 17 | IFNG | 1425947_at | 6018 | 0.234 | -0.0627 | No | ||
| 18 | NFKBIE | 1431843_a_at 1458299_s_at | 6075 | 0.232 | -0.0627 | No | ||
| 19 | PPP3R1 | 1421786_at 1433591_at 1450368_a_at | 6792 | 0.204 | -0.0933 | No | ||
| 20 | CBLC | 1422666_at 1431692_a_at | 7617 | 0.175 | -0.1291 | No | ||
| 21 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 9569 | 0.120 | -0.2171 | No | ||
| 22 | NFATC4 | 1423380_s_at 1432821_at 1454369_a_at | 10010 | 0.108 | -0.2361 | No | ||
| 23 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 10269 | 0.101 | -0.2468 | No | ||
| 24 | NFKBIB | 1421266_s_at 1446718_at | 10653 | 0.092 | -0.2633 | No | ||
| 25 | IL4 | 1449864_at | 10673 | 0.092 | -0.2632 | No | ||
| 26 | CSF2 | 1427429_at | 11291 | 0.076 | -0.2906 | No | ||
| 27 | CD3E | 1422105_at 1445748_at | 11496 | 0.071 | -0.2991 | No | ||
| 28 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 11615 | 0.067 | -0.3038 | No | ||
| 29 | TNF | 1419607_at | 11889 | 0.060 | -0.3157 | No | ||
| 30 | JUN | 1417409_at 1448694_at | 13314 | 0.024 | -0.3806 | No | ||
| 31 | PAK7 | 1440981_at 1446159_at 1455847_at 1458766_at | 13359 | 0.023 | -0.3823 | No | ||
| 32 | PAK3 | 1417923_at 1417924_at 1437318_at | 14187 | -0.003 | -0.4202 | No | ||
| 33 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 15150 | -0.039 | -0.4638 | No | ||
| 34 | PAK4 | 1428548_at | 15289 | -0.045 | -0.4696 | No | ||
| 35 | PIK3CB | 1445731_at 1453069_at | 15462 | -0.052 | -0.4769 | No | ||
| 36 | LAT | 1460651_at | 15852 | -0.072 | -0.4939 | No | ||
| 37 | PIK3CA | 1423144_at 1429434_at 1429435_x_at 1440054_at 1453134_at 1460326_at | 16083 | -0.086 | -0.5035 | No | ||
| 38 | CD40LG | 1422283_at | 16209 | -0.093 | -0.5082 | No | ||
| 39 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 16559 | -0.117 | -0.5229 | No | ||
| 40 | RHOA | 1437628_s_at | 17032 | -0.156 | -0.5428 | No | ||
| 41 | GRB2 | 1418508_a_at 1449111_a_at | 17171 | -0.170 | -0.5473 | No | ||
| 42 | AKT3 | 1422078_at 1435260_at 1435879_at 1443222_at 1460307_at | 17177 | -0.171 | -0.5457 | No | ||
| 43 | PAK6 | 1445475_at 1455200_at | 17358 | -0.192 | -0.5518 | No | ||
| 44 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 17435 | -0.202 | -0.5531 | No | ||
| 45 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 17487 | -0.209 | -0.5532 | No | ||
| 46 | NFKB2 | 1425902_a_at 1429128_x_at | 17945 | -0.270 | -0.5711 | No | ||
| 47 | MAP3K8 | 1419208_at | 18046 | -0.284 | -0.5726 | No | ||
| 48 | SOS2 | 1452281_at | 18613 | -0.379 | -0.5944 | No | ||
| 49 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 18818 | -0.421 | -0.5992 | Yes | ||
| 50 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 18829 | -0.424 | -0.5950 | Yes | ||
| 51 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 18974 | -0.459 | -0.5966 | Yes | ||
| 52 | SOS1 | 1421884_at 1421885_at 1421886_at | 18996 | -0.463 | -0.5925 | Yes | ||
| 53 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 19199 | -0.510 | -0.5962 | Yes | ||
| 54 | NFAT5 | 1426029_a_at 1438999_a_at 1439328_at 1439805_at 1448966_a_at 1451921_a_at 1458213_at | 19385 | -0.557 | -0.5986 | Yes | ||
| 55 | CTLA4 | 1419334_at | 19405 | -0.562 | -0.5934 | Yes | ||
| 56 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 19447 | -0.577 | -0.5890 | Yes | ||
| 57 | CD3D | 1422828_at | 19644 | -0.636 | -0.5910 | Yes | ||
| 58 | GRAP2 | 1456432_at | 19875 | -0.712 | -0.5938 | Yes | ||
| 59 | NRAS | 1422688_a_at 1454060_a_at | 19925 | -0.733 | -0.5880 | Yes | ||
| 60 | PTPN6 | 1456694_x_at 1460188_at | 20072 | -0.789 | -0.5861 | Yes | ||
| 61 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 20163 | -0.833 | -0.5812 | Yes | ||
| 62 | FOS | 1423100_at | 20289 | -0.888 | -0.5772 | Yes | ||
| 63 | IL5 | 1425546_a_at 1442049_at 1450550_at | 20299 | -0.893 | -0.5679 | Yes | ||
| 64 | ICOS | 1421930_at 1421931_at 1436598_at | 20392 | -0.937 | -0.5620 | Yes | ||
| 65 | VAV1 | 1422932_a_at | 20423 | -0.958 | -0.5529 | Yes | ||
| 66 | IKBKG | 1421208_at 1421209_s_at 1435646_at 1435647_at 1450161_at 1454690_at | 20443 | -0.967 | -0.5432 | Yes | ||
| 67 | CD8A | 1440164_x_at 1440811_x_at | 20574 | -1.039 | -0.5379 | Yes | ||
| 68 | PPP3CA | 1426401_at 1438478_a_at 1440051_at 1452056_s_at 1458495_at | 20582 | -1.046 | -0.5268 | Yes | ||
| 69 | CD3G | 1419178_at | 20668 | -1.109 | -0.5186 | Yes | ||
| 70 | PDK1 | 1423747_a_at 1423748_at 1434974_at 1435836_at | 20815 | -1.229 | -0.5119 | Yes | ||
| 71 | PRKCQ | 1426044_a_at | 20851 | -1.253 | -0.4999 | Yes | ||
| 72 | CBLB | 1437304_at 1455082_at 1458469_at | 20915 | -1.305 | -0.4886 | Yes | ||
| 73 | PAK2 | 1434250_at 1454887_at | 21124 | -1.581 | -0.4809 | Yes | ||
| 74 | LCP2 | 1418641_at 1418642_at | 21182 | -1.674 | -0.4653 | Yes | ||
| 75 | NFKB1 | 1427705_a_at 1442949_at | 21207 | -1.718 | -0.4477 | Yes | ||
| 76 | PPP3CB | 1427468_at 1428473_at 1428474_at 1433835_at 1446149_at 1459378_at | 21215 | -1.729 | -0.4292 | Yes | ||
| 77 | CARD11 | 1430257_at 1435996_at | 21299 | -1.844 | -0.4129 | Yes | ||
| 78 | PIK3R5 | 1434980_at | 21318 | -1.866 | -0.3934 | Yes | ||
| 79 | NFATC3 | 1419976_s_at 1452497_a_at | 21381 | -1.965 | -0.3749 | Yes | ||
| 80 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 21477 | -2.135 | -0.3560 | Yes | ||
| 81 | PIK3CD | 1422991_at 1422992_s_at 1443798_at 1453281_at 1458321_at | 21492 | -2.164 | -0.3331 | Yes | ||
| 82 | ITK | 1417171_at 1430833_at 1452518_a_at 1456836_at 1457120_at | 21504 | -2.182 | -0.3098 | Yes | ||
| 83 | CD28 | 1417597_at 1437025_at 1443703_at | 21558 | -2.314 | -0.2870 | Yes | ||
| 84 | CD4 | 1419696_at 1427779_a_at | 21629 | -2.582 | -0.2621 | Yes | ||
| 85 | PIK3CG | 1422707_at 1422708_at | 21647 | -2.651 | -0.2341 | Yes | ||
| 86 | RASGRP1 | 1421176_at 1431749_a_at 1434295_at 1450143_at | 21672 | -2.736 | -0.2054 | Yes | ||
| 87 | CD247 | 1420716_at 1426079_at 1426396_at 1428059_at 1428060_at 1438392_at 1452539_a_at | 21769 | -3.129 | -0.1757 | Yes | ||
| 88 | AKT2 | 1421324_a_at 1424480_s_at 1455703_at | 21832 | -3.540 | -0.1400 | Yes | ||
| 89 | VAV2 | 1421272_at 1435065_x_at 1435244_at 1458758_at | 21871 | -3.956 | -0.0987 | Yes | ||
| 90 | TEC | 1441524_at 1460204_at | 21881 | -4.130 | -0.0541 | Yes | ||
| 91 | CD8B | 1426170_a_at 1446372_at 1448569_at 1458652_at | 21914 | -5.200 | 0.0010 | Yes |