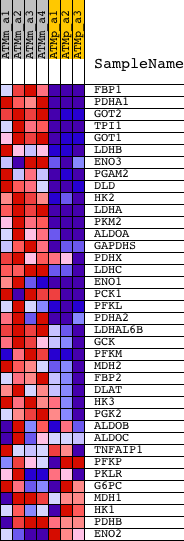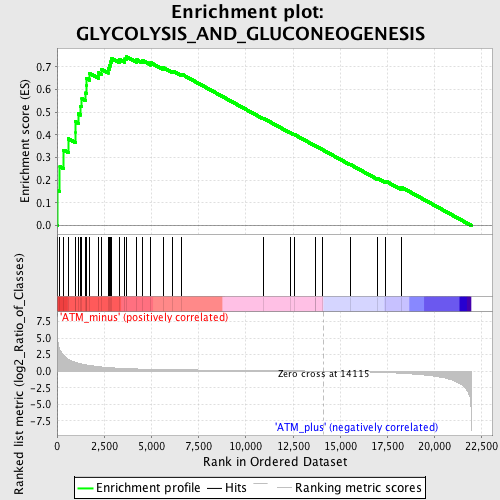
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | 0.74412423 |
| Normalized Enrichment Score (NES) | 1.9382871 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0011422267 |
| FWER p-Value | 0.01 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FBP1 | 1448470_at | 42 | 4.486 | 0.1541 | Yes | ||
| 2 | PDHA1 | 1418560_at 1446954_at 1449137_at | 150 | 3.164 | 0.2591 | Yes | ||
| 3 | GOT2 | 1417715_a_at 1417716_at | 348 | 2.373 | 0.3326 | Yes | ||
| 4 | TPI1 | 1415918_a_at 1435659_a_at 1452927_x_at | 606 | 1.765 | 0.3823 | Yes | ||
| 5 | GOT1 | 1450970_at | 968 | 1.333 | 0.4121 | Yes | ||
| 6 | LDHB | 1416183_a_at 1434499_a_at 1442618_at 1448237_x_at 1455235_x_at | 972 | 1.326 | 0.4581 | Yes | ||
| 7 | ENO3 | 1417951_at 1449631_at | 1128 | 1.196 | 0.4926 | Yes | ||
| 8 | PGAM2 | 1418373_at | 1242 | 1.117 | 0.5262 | Yes | ||
| 9 | DLD | 1423159_at | 1314 | 1.077 | 0.5604 | Yes | ||
| 10 | HK2 | 1422612_at | 1515 | 0.961 | 0.5847 | Yes | ||
| 11 | LDHA | 1419737_a_at | 1541 | 0.949 | 0.6166 | Yes | ||
| 12 | PKM2 | 1417308_at | 1548 | 0.945 | 0.6492 | Yes | ||
| 13 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 1722 | 0.867 | 0.6714 | Yes | ||
| 14 | GAPDHS | 1449974_at | 2192 | 0.685 | 0.6738 | Yes | ||
| 15 | PDHX | 1456090_at 1459434_at | 2352 | 0.636 | 0.6887 | Yes | ||
| 16 | LDHC | 1415846_a_at 1415847_at | 2702 | 0.548 | 0.6918 | Yes | ||
| 17 | ENO1 | 1419023_x_at | 2791 | 0.528 | 0.7061 | Yes | ||
| 18 | PCK1 | 1423439_at 1439617_s_at 1455209_at | 2847 | 0.518 | 0.7216 | Yes | ||
| 19 | PFKL | 1439148_a_at 1450269_a_at 1453541_at | 2888 | 0.510 | 0.7375 | Yes | ||
| 20 | PDHA2 | 1450962_at | 3301 | 0.437 | 0.7339 | Yes | ||
| 21 | LDHAL6B | 1434247_at | 3589 | 0.400 | 0.7347 | Yes | ||
| 22 | GCK | 1419146_a_at 1425303_at | 3680 | 0.389 | 0.7441 | Yes | ||
| 23 | PFKM | 1416780_at | 4224 | 0.335 | 0.7310 | No | ||
| 24 | MDH2 | 1416478_a_at 1433984_a_at | 4518 | 0.311 | 0.7284 | No | ||
| 25 | FBP2 | 1449088_at | 4966 | 0.284 | 0.7179 | No | ||
| 26 | DLAT | 1426264_at 1426265_x_at 1452005_at | 5622 | 0.253 | 0.6967 | No | ||
| 27 | HK3 | 1435490_at 1442798_x_at | 6132 | 0.230 | 0.6815 | No | ||
| 28 | PGK2 | 1416784_at | 6596 | 0.212 | 0.6677 | No | ||
| 29 | ALDOB | 1451194_at | 10909 | 0.086 | 0.4738 | No | ||
| 30 | ALDOC | 1424714_at 1451461_a_at | 12337 | 0.050 | 0.4104 | No | ||
| 31 | TNFAIP1 | 1417865_at 1417866_at 1448863_a_at | 12570 | 0.044 | 0.4013 | No | ||
| 32 | PFKP | 1416069_at 1430634_a_at 1437759_at | 13698 | 0.012 | 0.3503 | No | ||
| 33 | PKLR | 1421258_a_at 1421259_at 1438711_at | 13699 | 0.012 | 0.3507 | No | ||
| 34 | G6PC | 1417880_at | 14037 | 0.002 | 0.3354 | No | ||
| 35 | MDH1 | 1434319_at 1436834_x_at 1438338_at 1448172_at 1454925_x_at | 15522 | -0.055 | 0.2696 | No | ||
| 36 | HK1 | 1420901_a_at 1437974_a_at | 16986 | -0.152 | 0.2080 | No | ||
| 37 | PDHB | 1416090_at 1448214_at | 17392 | -0.196 | 0.1964 | No | ||
| 38 | ENO2 | 1418829_a_at | 18251 | -0.319 | 0.1683 | No |