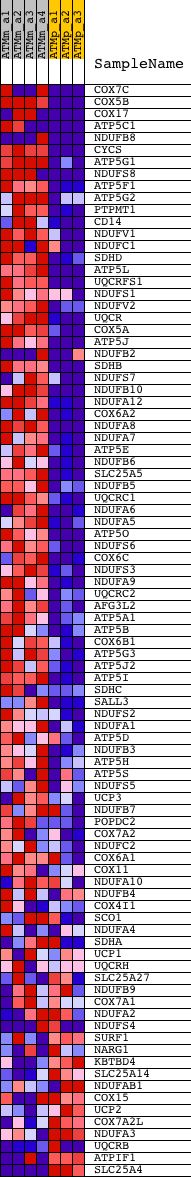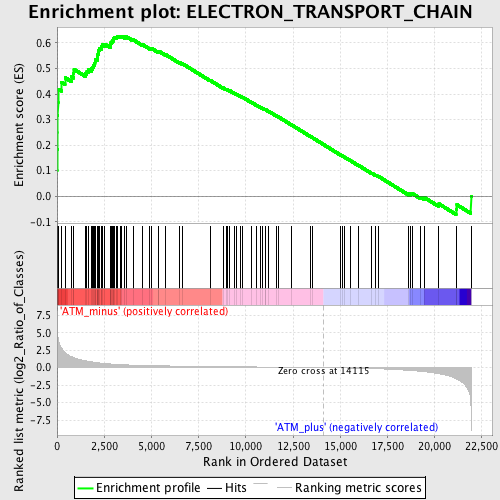
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | ELECTRON_TRANSPORT_CHAIN |
| Enrichment Score (ES) | 0.62652963 |
| Normalized Enrichment Score (NES) | 1.904286 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0029956074 |
| FWER p-Value | 0.032 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COX7C | 1448112_at 1459884_at 1459885_s_at | 0 | 8.097 | 0.0993 | Yes | ||
| 2 | COX5B | 1416902_a_at 1435613_x_at 1436149_at 1448514_at 1454716_x_at 1456588_x_at | 3 | 6.855 | 0.1833 | Yes | ||
| 3 | COX17 | 1434435_s_at 1434871_at | 13 | 5.555 | 0.2511 | Yes | ||
| 4 | ATP5C1 | 1416058_s_at 1438809_at | 15 | 5.347 | 0.3166 | Yes | ||
| 5 | NDUFB8 | 1448198_a_at 1458404_at | 46 | 4.285 | 0.3678 | Yes | ||
| 6 | CYCS | 1422484_at 1445484_at | 51 | 4.211 | 0.4193 | Yes | ||
| 7 | ATP5G1 | 1416020_a_at 1444874_at | 224 | 2.845 | 0.4463 | Yes | ||
| 8 | NDUFS8 | 1423907_a_at 1423908_at 1434212_at 1434213_x_at 1434579_x_at 1455283_x_at | 419 | 2.164 | 0.4640 | Yes | ||
| 9 | ATP5F1 | 1426742_at 1433562_s_at | 759 | 1.576 | 0.4678 | Yes | ||
| 10 | ATP5G2 | 1415980_at 1456128_at | 878 | 1.426 | 0.4799 | Yes | ||
| 11 | PTPMT1 | 1426581_at | 893 | 1.414 | 0.4966 | Yes | ||
| 12 | CD14 | 1417268_at | 1500 | 0.966 | 0.4807 | Yes | ||
| 13 | NDUFV1 | 1415966_a_at 1415967_at 1456015_x_at | 1579 | 0.931 | 0.4886 | Yes | ||
| 14 | NDUFC1 | 1416285_at 1448284_a_at | 1678 | 0.889 | 0.4950 | Yes | ||
| 15 | SDHD | 1428235_at 1437489_x_at | 1820 | 0.822 | 0.4986 | Yes | ||
| 16 | ATP5L | 1448203_at | 1892 | 0.789 | 0.5050 | Yes | ||
| 17 | UQCRFS1 | 1450968_at | 1931 | 0.774 | 0.5128 | Yes | ||
| 18 | NDUFS1 | 1425143_a_at 1453354_at | 1983 | 0.753 | 0.5197 | Yes | ||
| 19 | NDUFV2 | 1428179_at 1438159_x_at 1452692_a_at | 2007 | 0.743 | 0.5277 | Yes | ||
| 20 | UQCR | 1448292_at | 2018 | 0.740 | 0.5364 | Yes | ||
| 21 | COX5A | 1415933_a_at 1439267_x_at 1448153_at | 2132 | 0.703 | 0.5398 | Yes | ||
| 22 | ATP5J | 1416143_at | 2147 | 0.699 | 0.5478 | Yes | ||
| 23 | NDUFB2 | 1416834_x_at 1442658_at 1448483_a_at 1453806_at | 2158 | 0.695 | 0.5558 | Yes | ||
| 24 | SDHB | 1418005_at | 2180 | 0.690 | 0.5633 | Yes | ||
| 25 | NDUFS7 | 1424313_a_at 1451312_at | 2206 | 0.681 | 0.5705 | Yes | ||
| 26 | NDUFB10 | 1428322_a_at | 2233 | 0.673 | 0.5776 | Yes | ||
| 27 | NDUFA12 | 1418814_s_at 1433513_x_at | 2349 | 0.636 | 0.5801 | Yes | ||
| 28 | COX6A2 | 1417607_at 1457633_x_at | 2353 | 0.636 | 0.5878 | Yes | ||
| 29 | NDUFA8 | 1423692_at | 2386 | 0.627 | 0.5940 | Yes | ||
| 30 | NDUFA7 | 1422976_x_at 1428360_x_at 1436567_a_at 1450818_a_at 1455749_x_at | 2532 | 0.589 | 0.5946 | Yes | ||
| 31 | ATP5E | 1416567_s_at | 2817 | 0.525 | 0.5880 | Yes | ||
| 32 | NDUFB6 | 1434056_a_at 1434057_at | 2820 | 0.523 | 0.5944 | Yes | ||
| 33 | SLC25A5 | 1423772_x_at 1434801_x_at 1436874_x_at 1445944_at | 2842 | 0.519 | 0.5998 | Yes | ||
| 34 | NDUFB5 | 1417102_a_at 1448589_at | 2896 | 0.508 | 0.6036 | Yes | ||
| 35 | UQCRC1 | 1428782_a_at | 2915 | 0.503 | 0.6089 | Yes | ||
| 36 | NDUFA6 | 1448427_at | 2965 | 0.493 | 0.6127 | Yes | ||
| 37 | NDUFA5 | 1417285_a_at 1417286_at | 2983 | 0.491 | 0.6180 | Yes | ||
| 38 | ATP5O | 1416278_a_at | 3021 | 0.484 | 0.6222 | Yes | ||
| 39 | NDUFS6 | 1433603_at | 3153 | 0.461 | 0.6219 | Yes | ||
| 40 | COX6C | 1415970_at 1434491_a_at | 3175 | 0.457 | 0.6265 | Yes | ||
| 41 | NDUFS3 | 1423737_at | 3333 | 0.434 | 0.6247 | No | ||
| 42 | NDUFA9 | 1416663_at | 3425 | 0.420 | 0.6256 | No | ||
| 43 | UQCRC2 | 1428631_a_at 1435757_a_at | 3584 | 0.400 | 0.6233 | No | ||
| 44 | AFG3L2 | 1427206_at 1427207_s_at 1454003_at | 3678 | 0.389 | 0.6238 | No | ||
| 45 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 4020 | 0.352 | 0.6125 | No | ||
| 46 | ATP5B | 1416829_at | 4523 | 0.310 | 0.5934 | No | ||
| 47 | COX6B1 | 1416565_at 1436757_a_at | 4913 | 0.287 | 0.5791 | No | ||
| 48 | ATP5G3 | 1454661_at | 4990 | 0.283 | 0.5791 | No | ||
| 49 | ATP5J2 | 1416269_at 1435395_s_at | 5346 | 0.266 | 0.5661 | No | ||
| 50 | ATP5I | 1422525_at 1434053_x_at 1450640_x_at | 5377 | 0.264 | 0.5679 | No | ||
| 51 | SDHC | 1435986_x_at 1448630_a_at | 5740 | 0.247 | 0.5544 | No | ||
| 52 | SALL3 | 1441186_at 1452386_at | 6481 | 0.217 | 0.5232 | No | ||
| 53 | NDUFS2 | 1451096_at | 6654 | 0.210 | 0.5179 | No | ||
| 54 | NDUFA1 | 1422241_a_at | 8118 | 0.161 | 0.4529 | No | ||
| 55 | ATP5D | 1423716_s_at 1456580_s_at | 8787 | 0.141 | 0.4240 | No | ||
| 56 | NDUFB3 | 1416547_at | 8967 | 0.136 | 0.4175 | No | ||
| 57 | ATP5H | 1423676_at 1435112_a_at | 9029 | 0.135 | 0.4164 | No | ||
| 58 | ATP5S | 1417970_at 1459949_at | 9155 | 0.131 | 0.4123 | No | ||
| 59 | NDUFS5 | 1416494_at 1416495_s_at | 9379 | 0.125 | 0.4036 | No | ||
| 60 | UCP3 | 1420657_at 1420658_at | 9525 | 0.121 | 0.3984 | No | ||
| 61 | NDUFB7 | 1416417_a_at 1448331_at | 9696 | 0.117 | 0.3921 | No | ||
| 62 | POPDC2 | 1417806_at | 9814 | 0.113 | 0.3881 | No | ||
| 63 | COX7A2 | 1416970_a_at 1416971_at | 10291 | 0.101 | 0.3676 | No | ||
| 64 | NDUFC2 | 1431670_at | 10578 | 0.094 | 0.3556 | No | ||
| 65 | COX6A1 | 1417417_a_at | 10769 | 0.090 | 0.3480 | No | ||
| 66 | COX11 | 1429188_at 1457846_at | 10873 | 0.087 | 0.3444 | No | ||
| 67 | NDUFA10 | 1418068_at 1448934_at | 11014 | 0.083 | 0.3390 | No | ||
| 68 | NDUFB4 | 1428075_at 1428076_s_at 1432427_at | 11038 | 0.082 | 0.3389 | No | ||
| 69 | COX4I1 | 1448322_a_at | 11216 | 0.078 | 0.3318 | No | ||
| 70 | SCO1 | 1430300_at | 11625 | 0.067 | 0.3139 | No | ||
| 71 | NDUFA4 | 1424085_at | 11719 | 0.065 | 0.3105 | No | ||
| 72 | SDHA | 1426688_at 1426689_s_at 1445317_at | 12388 | 0.049 | 0.2805 | No | ||
| 73 | UCP1 | 1418197_at | 13419 | 0.021 | 0.2336 | No | ||
| 74 | UQCRH | 1452133_at 1453229_s_at | 13504 | 0.019 | 0.2300 | No | ||
| 75 | SLC25A27 | 1431201_at 1440090_at 1454230_a_at 1458119_at | 15024 | -0.034 | 0.1609 | No | ||
| 76 | NDUFB9 | 1436803_a_at 1452184_at 1455934_at | 15120 | -0.038 | 0.1570 | No | ||
| 77 | COX7A1 | 1418709_at | 15219 | -0.042 | 0.1530 | No | ||
| 78 | NDUFA2 | 1417368_s_at | 15514 | -0.055 | 0.1402 | No | ||
| 79 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 15987 | -0.080 | 0.1196 | No | ||
| 80 | SURF1 | 1450561_a_at | 16662 | -0.125 | 0.0903 | No | ||
| 81 | NARG1 | 1418022_at 1418023_at 1418024_at 1435139_at | 16880 | -0.143 | 0.0821 | No | ||
| 82 | KBTBD4 | 1417922_at | 17019 | -0.155 | 0.0777 | No | ||
| 83 | SLC25A14 | 1417154_at | 18620 | -0.380 | 0.0091 | No | ||
| 84 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 18725 | -0.400 | 0.0093 | No | ||
| 85 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 18815 | -0.421 | 0.0104 | No | ||
| 86 | UCP2 | 1448188_at 1459740_s_at 1459741_x_at | 19256 | -0.522 | -0.0034 | No | ||
| 87 | COX7A2L | 1421772_a_at 1432263_a_at 1432264_x_at | 19458 | -0.579 | -0.0055 | No | ||
| 88 | NDUFA3 | 1428464_at 1430149_at 1452790_x_at | 20225 | -0.856 | -0.0300 | No | ||
| 89 | UQCRB | 1416337_at 1455997_a_at | 21131 | -1.594 | -0.0519 | No | ||
| 90 | ATPIF1 | 1434198_at 1448770_a_at 1453681_at | 21175 | -1.665 | -0.0335 | No | ||
| 91 | SLC25A4 | 1424562_a_at 1424563_at 1434897_a_at 1455069_x_at | 21921 | -5.563 | 0.0007 | No |