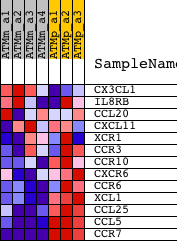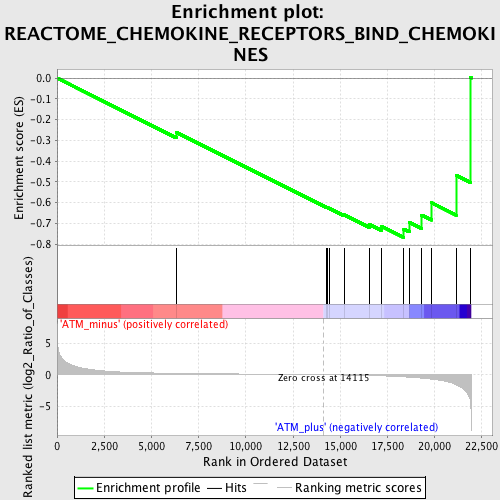
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | REACTOME_CHEMOKINE_RECEPTORS_BIND_CHEMOKINES |
| Enrichment Score (ES) | -0.76841646 |
| Normalized Enrichment Score (NES) | -1.6419797 |
| Nominal p-value | 0.012396694 |
| FDR q-value | 0.18391815 |
| FWER p-Value | 0.954 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CX3CL1 | 1415803_at 1415804_at 1421610_at 1441884_x_at | 6328 | 0.222 | -0.2620 | No | ||
| 2 | IL8RB | 1421734_at | 14257 | -0.005 | -0.6230 | No | ||
| 3 | CCL20 | 1422029_at | 14320 | -0.007 | -0.6250 | No | ||
| 4 | CXCL11 | 1419697_at 1419698_at | 14422 | -0.011 | -0.6283 | No | ||
| 5 | XCR1 | 1422294_at | 15226 | -0.042 | -0.6598 | No | ||
| 6 | CCR3 | 1422957_at | 16550 | -0.116 | -0.7063 | No | ||
| 7 | CCR10 | 1421420_at | 17179 | -0.171 | -0.7144 | No | ||
| 8 | CXCR6 | 1422812_at 1425832_a_at | 18364 | -0.336 | -0.7282 | Yes | ||
| 9 | CCR6 | 1450357_a_at | 18656 | -0.387 | -0.6951 | Yes | ||
| 10 | XCL1 | 1419412_at | 19294 | -0.534 | -0.6603 | Yes | ||
| 11 | CCL25 | 1418777_at 1458277_at | 19824 | -0.698 | -0.6009 | Yes | ||
| 12 | CCL5 | 1418126_at | 21135 | -1.602 | -0.4689 | Yes | ||
| 13 | CCR7 | 1423466_at | 21889 | -4.223 | 0.0021 | Yes |