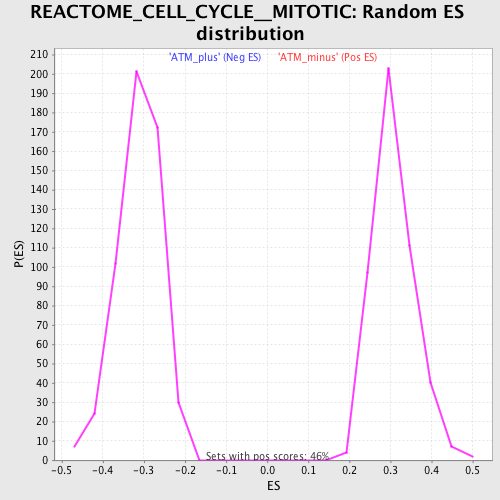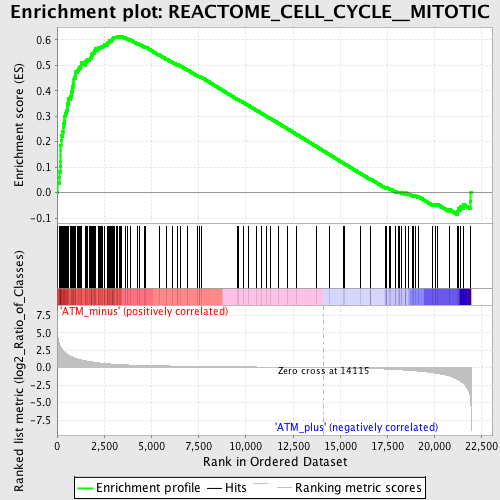
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_CELL_CYCLE__MITOTIC |
| Enrichment Score (ES) | 0.6146473 |
| Normalized Enrichment Score (NES) | 1.9960978 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.625E-4 |
| FWER p-Value | 0.0010 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 10 | 5.918 | 0.0411 | Yes | ||
| 2 | POLA2 | 1425794_at 1448369_at 1456713_at | 120 | 3.359 | 0.0596 | Yes | ||
| 3 | XPO1 | 1418442_at 1418443_at 1448070_at | 128 | 3.276 | 0.0823 | Yes | ||
| 4 | CCNB1 | 1419943_s_at 1419944_at | 168 | 3.093 | 0.1022 | Yes | ||
| 5 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 180 | 3.043 | 0.1231 | Yes | ||
| 6 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 184 | 3.018 | 0.1441 | Yes | ||
| 7 | PRIM1 | 1418369_at 1444544_at 1449061_a_at | 185 | 3.013 | 0.1653 | Yes | ||
| 8 | PSMC6 | 1417771_a_at | 188 | 2.991 | 0.1862 | Yes | ||
| 9 | POLE | 1441854_at 1448650_a_at | 208 | 2.910 | 0.2057 | Yes | ||
| 10 | ZWINT | 1423724_at 1427539_a_at 1427540_at 1429786_a_at 1429787_x_at 1444717_at 1455382_at | 236 | 2.731 | 0.2236 | Yes | ||
| 11 | ACTR1A | 1415873_a_at 1458696_at 1459855_x_at | 289 | 2.546 | 0.2391 | Yes | ||
| 12 | SDCCAG8 | 1431203_at 1444831_at 1453946_a_at 1456538_at | 323 | 2.444 | 0.2548 | Yes | ||
| 13 | FEN1 | 1421731_a_at 1436454_x_at | 341 | 2.391 | 0.2708 | Yes | ||
| 14 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 383 | 2.275 | 0.2848 | Yes | ||
| 15 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 401 | 2.214 | 0.2996 | Yes | ||
| 16 | CDKN1A | 1421679_a_at 1424638_at | 454 | 2.078 | 0.3118 | Yes | ||
| 17 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 517 | 1.943 | 0.3226 | Yes | ||
| 18 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 531 | 1.905 | 0.3354 | Yes | ||
| 19 | NEK2 | 1417299_at 1437579_at 1437580_s_at 1443999_at | 535 | 1.890 | 0.3485 | Yes | ||
| 20 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 584 | 1.808 | 0.3590 | Yes | ||
| 21 | CENPA | 1441864_x_at 1444416_at 1450842_a_at | 599 | 1.779 | 0.3708 | Yes | ||
| 22 | SMC1A | 1417830_at 1417831_at 1417832_at | 718 | 1.620 | 0.3768 | Yes | ||
| 23 | PSMD7 | 1432820_at 1451056_at 1454368_at | 758 | 1.577 | 0.3860 | Yes | ||
| 24 | RFC4 | 1424321_at 1438161_s_at | 762 | 1.572 | 0.3969 | Yes | ||
| 25 | MCM5 | 1415945_at 1436808_x_at | 806 | 1.510 | 0.4056 | Yes | ||
| 26 | WEE1 | 1416773_at 1416774_at | 812 | 1.497 | 0.4158 | Yes | ||
| 27 | RFC3 | 1423700_at 1432538_a_at | 843 | 1.469 | 0.4248 | Yes | ||
| 28 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 888 | 1.419 | 0.4327 | Yes | ||
| 29 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 891 | 1.416 | 0.4426 | Yes | ||
| 30 | MCM4 | 1416214_at 1436708_x_at | 896 | 1.412 | 0.4523 | Yes | ||
| 31 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 973 | 1.326 | 0.4581 | Yes | ||
| 32 | POLE2 | 1427094_at 1431440_at | 991 | 1.311 | 0.4665 | Yes | ||
| 33 | PSMD12 | 1448492_a_at | 993 | 1.310 | 0.4757 | Yes | ||
| 34 | PSMA4 | 1460339_at | 1094 | 1.216 | 0.4796 | Yes | ||
| 35 | NUP160 | 1418530_at 1444892_at | 1131 | 1.194 | 0.4863 | Yes | ||
| 36 | NUP43 | 1432187_at 1432188_s_at | 1168 | 1.170 | 0.4929 | Yes | ||
| 37 | PSME3 | 1418078_at 1418079_at 1438509_at | 1254 | 1.110 | 0.4968 | Yes | ||
| 38 | GMNN | 1417506_at | 1266 | 1.104 | 0.5040 | Yes | ||
| 39 | POLD2 | 1448277_at | 1306 | 1.081 | 0.5098 | Yes | ||
| 40 | CDK4 | 1422441_x_at | 1506 | 0.963 | 0.5074 | Yes | ||
| 41 | BIRC5 | 1424278_a_at | 1508 | 0.963 | 0.5142 | Yes | ||
| 42 | KIF20A | 1449207_a_at | 1560 | 0.939 | 0.5184 | Yes | ||
| 43 | LIG1 | 1416641_at | 1607 | 0.917 | 0.5227 | Yes | ||
| 44 | BUB1 | 1424046_at 1438571_at | 1704 | 0.876 | 0.5245 | Yes | ||
| 45 | MAD2L1 | 1422460_at | 1742 | 0.858 | 0.5288 | Yes | ||
| 46 | PSMA2 | 1448206_at | 1800 | 0.830 | 0.5320 | Yes | ||
| 47 | PSMC1 | 1416005_at | 1826 | 0.820 | 0.5366 | Yes | ||
| 48 | PSMB10 | 1448632_at | 1832 | 0.817 | 0.5421 | Yes | ||
| 49 | RPA2 | 1416433_at 1454011_a_at | 1887 | 0.793 | 0.5452 | Yes | ||
| 50 | RFC5 | 1452917_at | 1900 | 0.786 | 0.5502 | Yes | ||
| 51 | PSMC4 | 1416290_a_at 1416291_at | 1975 | 0.757 | 0.5521 | Yes | ||
| 52 | PSMA7 | 1423567_a_at 1423568_at | 1978 | 0.756 | 0.5573 | Yes | ||
| 53 | PSMA5 | 1424681_a_at 1434356_a_at | 2010 | 0.742 | 0.5611 | Yes | ||
| 54 | POLD1 | 1448187_at 1456055_x_at | 2046 | 0.729 | 0.5646 | Yes | ||
| 55 | PMF1 | 1416707_a_at 1438173_x_at | 2167 | 0.693 | 0.5639 | Yes | ||
| 56 | CDC25C | 1422252_a_at | 2169 | 0.693 | 0.5688 | Yes | ||
| 57 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 2251 | 0.668 | 0.5697 | Yes | ||
| 58 | PSMB9 | 1450696_at | 2314 | 0.646 | 0.5714 | Yes | ||
| 59 | POLD3 | 1426838_at 1426839_at 1443733_x_at 1459647_at | 2337 | 0.640 | 0.5749 | Yes | ||
| 60 | CENPE | 1435005_at 1439040_at | 2414 | 0.620 | 0.5758 | Yes | ||
| 61 | TUBG1 | 1417144_at | 2486 | 0.599 | 0.5767 | Yes | ||
| 62 | PSMC3 | 1416282_at | 2493 | 0.597 | 0.5806 | Yes | ||
| 63 | KNTC1 | 1435575_at | 2533 | 0.588 | 0.5830 | Yes | ||
| 64 | PSMD6 | 1423696_a_at 1423697_at | 2647 | 0.563 | 0.5817 | Yes | ||
| 65 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 2670 | 0.558 | 0.5846 | Yes | ||
| 66 | PSMC5 | 1415740_at | 2684 | 0.553 | 0.5879 | Yes | ||
| 67 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 2708 | 0.547 | 0.5907 | Yes | ||
| 68 | CEP135 | 1444201_at | 2750 | 0.536 | 0.5926 | Yes | ||
| 69 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 2752 | 0.535 | 0.5963 | Yes | ||
| 70 | NUP37 | 1423969_at | 2807 | 0.526 | 0.5975 | Yes | ||
| 71 | PSMD5 | 1423234_at | 2894 | 0.509 | 0.5971 | Yes | ||
| 72 | PSMD8 | 1423296_at | 2913 | 0.504 | 0.5998 | Yes | ||
| 73 | PSMA3 | 1448442_a_at | 2943 | 0.497 | 0.6020 | Yes | ||
| 74 | CCNE1 | 1416492_at 1441910_x_at | 2955 | 0.495 | 0.6050 | Yes | ||
| 75 | PLK4 | 1419838_s_at 1426580_at 1452115_a_at | 2973 | 0.492 | 0.6076 | Yes | ||
| 76 | PCNA | 1417947_at | 2980 | 0.492 | 0.6108 | Yes | ||
| 77 | PSMB5 | 1415676_a_at | 3059 | 0.476 | 0.6106 | Yes | ||
| 78 | CEP57 | 1428968_at 1452983_at | 3129 | 0.464 | 0.6107 | Yes | ||
| 79 | FGFR1OP | 1428919_at | 3180 | 0.456 | 0.6116 | Yes | ||
| 80 | ORC1L | 1422663_at 1443172_at | 3218 | 0.449 | 0.6130 | Yes | ||
| 81 | PSMD3 | 1448479_at | 3314 | 0.436 | 0.6117 | Yes | ||
| 82 | CENPC1 | 1420441_at | 3355 | 0.430 | 0.6129 | Yes | ||
| 83 | MCM2 | 1434079_s_at 1448777_at | 3383 | 0.426 | 0.6146 | Yes | ||
| 84 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 3596 | 0.399 | 0.6077 | No | ||
| 85 | DCTN3 | 1416247_at | 3721 | 0.385 | 0.6047 | No | ||
| 86 | TYMS | 1427810_at 1427811_at 1438690_at 1442192_at | 3867 | 0.368 | 0.6007 | No | ||
| 87 | CCNH | 1418585_at | 4269 | 0.332 | 0.5846 | No | ||
| 88 | UBE2C | 1452954_at | 4374 | 0.323 | 0.5821 | No | ||
| 89 | CDC27 | 1426076_at 1433848_at 1433849_at 1454739_at | 4618 | 0.304 | 0.5730 | No | ||
| 90 | PSMB4 | 1438984_x_at 1451205_at | 4692 | 0.300 | 0.5718 | No | ||
| 91 | PRKAR2B | 1430640_a_at 1438664_at 1456475_s_at | 5402 | 0.263 | 0.5411 | No | ||
| 92 | OFD1 | 1427172_at | 5813 | 0.244 | 0.5240 | No | ||
| 93 | NUP107 | 1426751_s_at | 6103 | 0.230 | 0.5123 | No | ||
| 94 | SGOL2 | 1437370_at | 6349 | 0.222 | 0.5027 | No | ||
| 95 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 6368 | 0.221 | 0.5034 | No | ||
| 96 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 6516 | 0.215 | 0.4981 | No | ||
| 97 | CCNB2 | 1450920_at | 6922 | 0.199 | 0.4809 | No | ||
| 98 | ORC5L | 1415830_at 1441601_at | 7409 | 0.182 | 0.4599 | No | ||
| 99 | KIF2A | 1432072_at 1450052_at 1450053_at 1452499_a_at 1454107_a_at | 7537 | 0.177 | 0.4553 | No | ||
| 100 | DYNLL1 | 1448682_at | 7543 | 0.177 | 0.4563 | No | ||
| 101 | POLA1 | 1419397_at 1456285_at 1459319_at 1459963_at | 7660 | 0.174 | 0.4522 | No | ||
| 102 | CDC16 | 1425554_a_at | 9545 | 0.121 | 0.3666 | No | ||
| 103 | MNAT1 | 1432177_a_at | 9602 | 0.119 | 0.3649 | No | ||
| 104 | PSMD2 | 1415831_at | 9867 | 0.112 | 0.3535 | No | ||
| 105 | CEP110 | 1421004_at 1421005_at 1435779_at | 10161 | 0.104 | 0.3408 | No | ||
| 106 | CEP152 | 1427496_at | 10584 | 0.094 | 0.3221 | No | ||
| 107 | CEP78 | 1436352_at | 10847 | 0.087 | 0.3107 | No | ||
| 108 | CKAP5 | 1430647_at 1437349_at 1442271_at | 11104 | 0.081 | 0.2995 | No | ||
| 109 | DCTN2 | 1424461_at | 11312 | 0.075 | 0.2905 | No | ||
| 110 | SSNA1 | 1448643_at | 11725 | 0.065 | 0.2721 | No | ||
| 111 | TUBB4 | 1423221_at | 12203 | 0.053 | 0.2506 | No | ||
| 112 | CETN2 | 1418579_at 1438647_x_at | 12689 | 0.041 | 0.2286 | No | ||
| 113 | CDC25B | 1421963_a_at | 13756 | 0.011 | 0.1797 | No | ||
| 114 | CCNA1 | 1449177_at | 14444 | -0.012 | 0.1483 | No | ||
| 115 | CUL1 | 1448174_at 1459516_at | 15156 | -0.039 | 0.1159 | No | ||
| 116 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 15208 | -0.041 | 0.1139 | No | ||
| 117 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 16069 | -0.085 | 0.0750 | No | ||
| 118 | RFC1 | 1418342_at 1449050_at 1451920_a_at | 16578 | -0.118 | 0.0525 | No | ||
| 119 | PRKACA | 1447720_x_at 1450519_a_at | 16584 | -0.119 | 0.0531 | No | ||
| 120 | PLK1 | 1443408_at 1448191_at 1459616_at | 16609 | -0.121 | 0.0529 | No | ||
| 121 | ORC2L | 1418225_at 1418226_at 1418227_at | 17417 | -0.200 | 0.0172 | No | ||
| 122 | ZW10 | 1424891_a_at 1431747_at 1445206_at | 17420 | -0.200 | 0.0185 | No | ||
| 123 | STAG2 | 1421849_at 1450396_at | 17433 | -0.202 | 0.0194 | No | ||
| 124 | RAD21 | 1416161_at 1416162_at 1455938_x_at | 17460 | -0.205 | 0.0196 | No | ||
| 125 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 17624 | -0.229 | 0.0138 | No | ||
| 126 | CDC25A | 1417131_at 1417132_at 1445995_at | 17656 | -0.233 | 0.0140 | No | ||
| 127 | RANBP2 | 1422621_at 1440104_at 1445883_at 1450690_at | 17929 | -0.269 | 0.0034 | No | ||
| 128 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 17944 | -0.270 | 0.0046 | No | ||
| 129 | CLASP2 | 1424836_a_at 1427328_a_at 1429976_at 1440802_at 1441726_at 1443320_at 1456911_at | 18062 | -0.287 | 0.0013 | No | ||
| 130 | NUMA1 | 1423974_at 1423975_s_at 1441090_at 1444576_at 1444577_x_at | 18114 | -0.296 | 0.0010 | No | ||
| 131 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18142 | -0.301 | 0.0019 | No | ||
| 132 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 18242 | -0.317 | -0.0004 | No | ||
| 133 | SFI1 | 1428778_at | 18252 | -0.319 | 0.0014 | No | ||
| 134 | CDK2 | 1416873_a_at 1447617_at | 18435 | -0.345 | -0.0045 | No | ||
| 135 | CCNA2 | 1417910_at 1417911_at | 18439 | -0.346 | -0.0023 | No | ||
| 136 | RPA3 | 1448938_at | 18476 | -0.353 | -0.0014 | No | ||
| 137 | TK2 | 1426100_a_at | 18633 | -0.384 | -0.0059 | No | ||
| 138 | TAOK1 | 1424657_at 1424658_at 1426357_at 1426358_at 1444385_at 1455432_at 1456975_at 1459770_at | 18843 | -0.427 | -0.0125 | No | ||
| 139 | DYNC1I2 | 1415841_at | 18876 | -0.435 | -0.0109 | No | ||
| 140 | KIF23 | 1450827_at 1453748_a_at 1455990_at | 18961 | -0.456 | -0.0116 | No | ||
| 141 | TUBB2C | 1423642_at 1438622_x_at 1439416_x_at 1456031_at 1456078_x_at 1456470_x_at | 19161 | -0.503 | -0.0172 | No | ||
| 142 | DHFR | 1419172_at 1430750_at | 19889 | -0.717 | -0.0455 | No | ||
| 143 | CDC45L | 1416575_at 1457838_at | 20026 | -0.772 | -0.0463 | No | ||
| 144 | SGOL1 | 1418919_at 1439510_at | 20121 | -0.813 | -0.0450 | No | ||
| 145 | UBE2D1 | 1424062_at | 20758 | -1.180 | -0.0659 | No | ||
| 146 | PPP1CC | 1440594_at | 21199 | -1.702 | -0.0741 | No | ||
| 147 | CSNK1E | 1417175_at 1417176_at 1435373_at | 21233 | -1.757 | -0.0633 | No | ||
| 148 | MCM3 | 1420029_at 1426653_at | 21359 | -1.933 | -0.0555 | No | ||
| 149 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 21536 | -2.261 | -0.0477 | No | ||
| 150 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 21877 | -4.048 | -0.0349 | No | ||
| 151 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 21918 | -5.349 | 0.0008 | No |