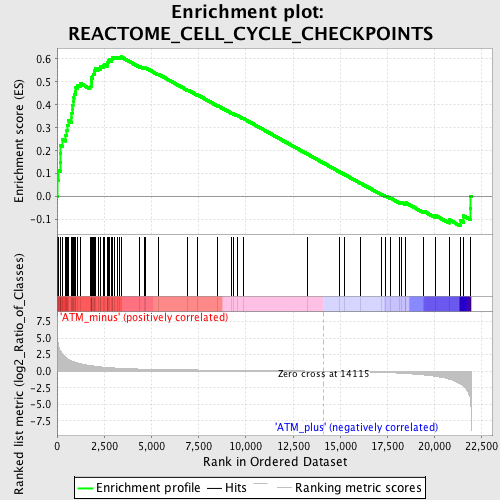
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_CELL_CYCLE_CHECKPOINTS |
| Enrichment Score (ES) | 0.6091298 |
| Normalized Enrichment Score (NES) | 1.7813356 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.020086246 |
| FWER p-Value | 0.265 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 10 | 5.918 | 0.0726 | Yes | ||
| 2 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 82 | 3.710 | 0.1152 | Yes | ||
| 3 | CCNB1 | 1419943_s_at 1419944_at | 168 | 3.093 | 0.1495 | Yes | ||
| 4 | PSMD9 | 1423386_at 1423387_at 1444321_at 1447670_at | 180 | 3.043 | 0.1866 | Yes | ||
| 5 | PSMC6 | 1417771_a_at | 188 | 2.991 | 0.2232 | Yes | ||
| 6 | MCM6 | 1416251_at 1438852_x_at 1447756_x_at | 293 | 2.536 | 0.2497 | Yes | ||
| 7 | CDKN1A | 1421679_a_at 1424638_at | 454 | 2.078 | 0.2681 | Yes | ||
| 8 | MCM10 | 1433407_at 1433408_a_at 1454244_at | 517 | 1.943 | 0.2892 | Yes | ||
| 9 | BUB1B | 1416961_at 1447362_at 1447363_s_at | 531 | 1.905 | 0.3121 | Yes | ||
| 10 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 584 | 1.808 | 0.3321 | Yes | ||
| 11 | PSMD7 | 1432820_at 1451056_at 1454368_at | 758 | 1.577 | 0.3437 | Yes | ||
| 12 | RFC4 | 1424321_at 1438161_s_at | 762 | 1.572 | 0.3629 | Yes | ||
| 13 | MCM5 | 1415945_at 1436808_x_at | 806 | 1.510 | 0.3796 | Yes | ||
| 14 | WEE1 | 1416773_at 1416774_at | 812 | 1.497 | 0.3979 | Yes | ||
| 15 | RFC3 | 1423700_at 1432538_a_at | 843 | 1.469 | 0.4146 | Yes | ||
| 16 | ORC4L | 1423336_at 1423337_at 1431172_at 1453804_a_at | 888 | 1.419 | 0.4301 | Yes | ||
| 17 | MCM4 | 1416214_at 1436708_x_at | 896 | 1.412 | 0.4473 | Yes | ||
| 18 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 973 | 1.326 | 0.4602 | Yes | ||
| 19 | PSMD12 | 1448492_a_at | 993 | 1.310 | 0.4755 | Yes | ||
| 20 | PSMA4 | 1460339_at | 1094 | 1.216 | 0.4859 | Yes | ||
| 21 | PSME3 | 1418078_at 1418079_at 1438509_at | 1254 | 1.110 | 0.4923 | Yes | ||
| 22 | MAD2L1 | 1422460_at | 1742 | 0.858 | 0.4807 | Yes | ||
| 23 | PSMA2 | 1448206_at | 1800 | 0.830 | 0.4883 | Yes | ||
| 24 | PSMB3 | 1417052_at | 1802 | 0.829 | 0.4985 | Yes | ||
| 25 | PSMC1 | 1416005_at | 1826 | 0.820 | 0.5076 | Yes | ||
| 26 | PSMB10 | 1448632_at | 1832 | 0.817 | 0.5174 | Yes | ||
| 27 | RPA2 | 1416433_at 1454011_a_at | 1887 | 0.793 | 0.5247 | Yes | ||
| 28 | RFC5 | 1452917_at | 1900 | 0.786 | 0.5339 | Yes | ||
| 29 | PSMC4 | 1416290_a_at 1416291_at | 1975 | 0.757 | 0.5399 | Yes | ||
| 30 | PSMA7 | 1423567_a_at 1423568_at | 1978 | 0.756 | 0.5491 | Yes | ||
| 31 | PSMA5 | 1424681_a_at 1434356_a_at | 2010 | 0.742 | 0.5568 | Yes | ||
| 32 | CDC25C | 1422252_a_at | 2169 | 0.693 | 0.5582 | Yes | ||
| 33 | CHEK1 | 1420031_at 1420032_at 1439208_at 1449708_s_at 1450677_at | 2290 | 0.655 | 0.5608 | Yes | ||
| 34 | PSMB9 | 1450696_at | 2314 | 0.646 | 0.5677 | Yes | ||
| 35 | PSMB6 | 1448822_at | 2439 | 0.614 | 0.5696 | Yes | ||
| 36 | PSMC3 | 1416282_at | 2493 | 0.597 | 0.5746 | Yes | ||
| 37 | PSMD6 | 1423696_a_at 1423697_at | 2647 | 0.563 | 0.5745 | Yes | ||
| 38 | PSMB1 | 1420052_x_at 1420053_at 1448166_a_at | 2670 | 0.558 | 0.5804 | Yes | ||
| 39 | PSMC5 | 1415740_at | 2684 | 0.553 | 0.5866 | Yes | ||
| 40 | MCM7 | 1416030_a_at 1416031_s_at 1438320_s_at | 2708 | 0.547 | 0.5923 | Yes | ||
| 41 | RPA1 | 1423293_at 1437309_a_at 1441240_at | 2752 | 0.535 | 0.5970 | Yes | ||
| 42 | PSMD5 | 1423234_at | 2894 | 0.509 | 0.5968 | Yes | ||
| 43 | PSMD8 | 1423296_at | 2913 | 0.504 | 0.6022 | Yes | ||
| 44 | PSMA3 | 1448442_a_at | 2943 | 0.497 | 0.6070 | Yes | ||
| 45 | PSMB5 | 1415676_a_at | 3059 | 0.476 | 0.6076 | Yes | ||
| 46 | ORC1L | 1422663_at 1443172_at | 3218 | 0.449 | 0.6059 | Yes | ||
| 47 | PSMD3 | 1448479_at | 3314 | 0.436 | 0.6070 | Yes | ||
| 48 | MCM2 | 1434079_s_at 1448777_at | 3383 | 0.426 | 0.6091 | Yes | ||
| 49 | UBE2C | 1452954_at | 4374 | 0.323 | 0.5678 | No | ||
| 50 | CDC27 | 1426076_at 1433848_at 1433849_at 1454739_at | 4618 | 0.304 | 0.5605 | No | ||
| 51 | PSMB4 | 1438984_x_at 1451205_at | 4692 | 0.300 | 0.5608 | No | ||
| 52 | RAD1 | 1448414_at | 5365 | 0.265 | 0.5334 | No | ||
| 53 | CCNB2 | 1450920_at | 6922 | 0.199 | 0.4646 | No | ||
| 54 | ORC5L | 1415830_at 1441601_at | 7409 | 0.182 | 0.4447 | No | ||
| 55 | CDC7 | 1426002_a_at 1426021_a_at | 8519 | 0.149 | 0.3958 | No | ||
| 56 | PSME1 | 1417056_at | 9239 | 0.129 | 0.3645 | No | ||
| 57 | RAD9B | 1425800_at 1439955_at | 9355 | 0.126 | 0.3608 | No | ||
| 58 | CDC16 | 1425554_a_at | 9545 | 0.121 | 0.3536 | No | ||
| 59 | PSMD2 | 1415831_at | 9867 | 0.112 | 0.3403 | No | ||
| 60 | HUS1 | 1418308_at 1425366_a_at | 13265 | 0.026 | 0.1852 | No | ||
| 61 | ATM | 1421205_at 1428830_at | 14971 | -0.032 | 0.1076 | No | ||
| 62 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 15208 | -0.041 | 0.0973 | No | ||
| 63 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 16069 | -0.085 | 0.0590 | No | ||
| 64 | PSMD10 | 1436559_a_at | 17175 | -0.171 | 0.0106 | No | ||
| 65 | ORC2L | 1418225_at 1418226_at 1418227_at | 17417 | -0.200 | 0.0020 | No | ||
| 66 | CDC25A | 1417131_at 1417132_at 1445995_at | 17656 | -0.233 | -0.0060 | No | ||
| 67 | PSMB2 | 1444377_at 1448262_at 1459641_at | 18142 | -0.301 | -0.0245 | No | ||
| 68 | RFC2 | 1417503_at 1457638_x_at 1457669_x_at | 18242 | -0.317 | -0.0251 | No | ||
| 69 | CDK2 | 1416873_a_at 1447617_at | 18435 | -0.345 | -0.0296 | No | ||
| 70 | RPA3 | 1448938_at | 18476 | -0.353 | -0.0271 | No | ||
| 71 | MDM2 | 1423605_a_at 1427718_a_at 1457929_at | 19427 | -0.568 | -0.0635 | No | ||
| 72 | CDC45L | 1416575_at 1457838_at | 20026 | -0.772 | -0.0813 | No | ||
| 73 | UBE2D1 | 1424062_at | 20758 | -1.180 | -0.1002 | No | ||
| 74 | MCM3 | 1420029_at 1426653_at | 21359 | -1.933 | -0.1038 | No | ||
| 75 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 21536 | -2.261 | -0.0839 | No | ||
| 76 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 21877 | -4.048 | -0.0495 | No | ||
| 77 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21888 | -4.222 | 0.0022 | No |

