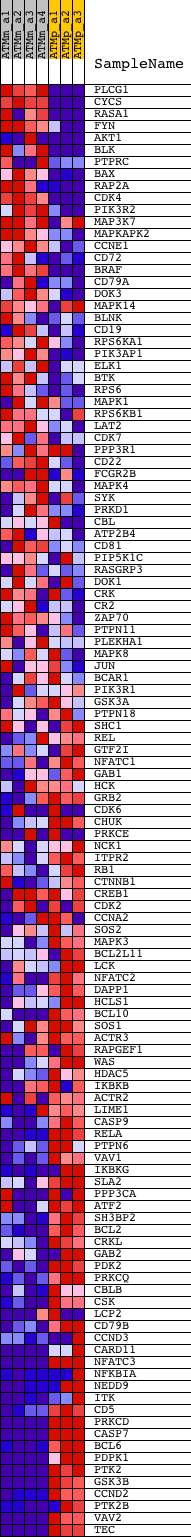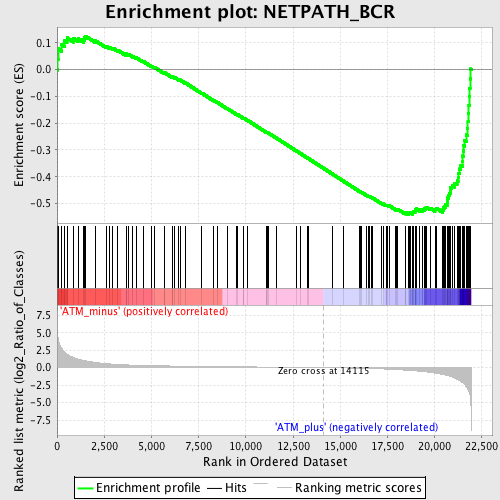
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NETPATH_BCR |
| Enrichment Score (ES) | -0.540929 |
| Normalized Enrichment Score (NES) | -1.6644558 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18632911 |
| FWER p-Value | 0.911 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PLCG1 | 1435149_at 1450360_at | 37 | 4.528 | 0.0396 | No | ||
| 2 | CYCS | 1422484_at 1445484_at | 51 | 4.211 | 0.0774 | No | ||
| 3 | RASA1 | 1426476_at 1426477_at 1426478_at 1438998_at | 243 | 2.703 | 0.0933 | No | ||
| 4 | FYN | 1417558_at 1441647_at 1448765_at | 384 | 2.271 | 0.1076 | No | ||
| 5 | AKT1 | 1416657_at 1425711_a_at 1440950_at 1442759_at | 525 | 1.916 | 0.1187 | No | ||
| 6 | BLK | 1422775_at | 869 | 1.437 | 0.1161 | No | ||
| 7 | PTPRC | 1422124_a_at 1440165_at | 1114 | 1.204 | 0.1159 | No | ||
| 8 | BAX | 1416837_at | 1403 | 1.015 | 0.1119 | No | ||
| 9 | RAP2A | 1421574_at 1426965_at 1440473_at | 1450 | 0.991 | 0.1189 | No | ||
| 10 | CDK4 | 1422441_x_at | 1506 | 0.963 | 0.1251 | No | ||
| 11 | PIK3R2 | 1418463_at | 2025 | 0.739 | 0.1081 | No | ||
| 12 | MAP3K7 | 1419988_at 1425795_a_at 1426627_at 1449693_at 1455441_at 1458787_at | 2595 | 0.574 | 0.0873 | No | ||
| 13 | MAPKAPK2 | 1426648_at | 2765 | 0.533 | 0.0844 | No | ||
| 14 | CCNE1 | 1416492_at 1441910_x_at | 2955 | 0.495 | 0.0802 | No | ||
| 15 | CD72 | 1426112_a_at | 3223 | 0.448 | 0.0721 | No | ||
| 16 | BRAF | 1425693_at 1435434_at 1435480_at 1442749_at 1445786_at 1447940_a_at 1447941_x_at 1456505_at 1458641_at | 3656 | 0.391 | 0.0559 | No | ||
| 17 | CD79A | 1418830_at | 3670 | 0.389 | 0.0588 | No | ||
| 18 | DOK3 | 1418096_at | 3773 | 0.379 | 0.0576 | No | ||
| 19 | MAPK14 | 1416703_at 1416704_at 1426104_at 1442364_at 1451927_a_at 1459617_at | 4014 | 0.353 | 0.0498 | No | ||
| 20 | BLNK | 1451780_at | 4193 | 0.337 | 0.0447 | No | ||
| 21 | CD19 | 1450570_a_at | 4551 | 0.309 | 0.0312 | No | ||
| 22 | RPS6KA1 | 1416896_at | 5001 | 0.282 | 0.0132 | No | ||
| 23 | PIK3AP1 | 1421285_at 1429831_at | 5155 | 0.275 | 0.0087 | No | ||
| 24 | ELK1 | 1421896_at 1421897_at 1446390_at | 5676 | 0.250 | -0.0129 | No | ||
| 25 | BTK | 1422755_at | 5693 | 0.249 | -0.0113 | No | ||
| 26 | RPS6 | 1453466_at | 6085 | 0.231 | -0.0272 | No | ||
| 27 | MAPK1 | 1419568_at 1426585_s_at 1442876_at 1453104_at | 6119 | 0.230 | -0.0266 | No | ||
| 28 | RPS6KB1 | 1428849_at 1446376_at 1454956_at 1457562_at 1459951_at 1460705_at | 6240 | 0.226 | -0.0300 | No | ||
| 29 | LAT2 | 1426169_a_at | 6453 | 0.218 | -0.0377 | No | ||
| 30 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 6516 | 0.215 | -0.0386 | No | ||
| 31 | PPP3R1 | 1421786_at 1433591_at 1450368_a_at | 6792 | 0.204 | -0.0494 | No | ||
| 32 | CD22 | 1419768_at 1419769_at 1445222_at | 7656 | 0.174 | -0.0873 | No | ||
| 33 | FCGR2B | 1435476_a_at 1435477_s_at 1447622_at 1451941_a_at 1455332_x_at | 8308 | 0.155 | -0.1158 | No | ||
| 34 | MAPK4 | 1435367_at | 8479 | 0.150 | -0.1222 | No | ||
| 35 | SYK | 1418261_at 1418262_at 1425797_a_at 1457239_at | 9007 | 0.135 | -0.1451 | No | ||
| 36 | PRKD1 | 1422673_at | 9487 | 0.122 | -0.1660 | No | ||
| 37 | CBL | 1434829_at 1446608_at 1450457_at 1455886_at | 9569 | 0.120 | -0.1686 | No | ||
| 38 | ATP2B4 | 1445701_at | 9889 | 0.111 | -0.1822 | No | ||
| 39 | CD81 | 1416330_at | 9895 | 0.111 | -0.1814 | No | ||
| 40 | PIP5K1C | 1450228_a_at | 10077 | 0.106 | -0.1887 | No | ||
| 41 | RASGRP3 | 1438030_at 1438031_at | 11086 | 0.081 | -0.2342 | No | ||
| 42 | DOK1 | 1417790_at | 11099 | 0.081 | -0.2340 | No | ||
| 43 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 11135 | 0.080 | -0.2349 | No | ||
| 44 | CR2 | 1425289_a_at | 11217 | 0.078 | -0.2379 | No | ||
| 45 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 11615 | 0.067 | -0.2555 | No | ||
| 46 | PTPN11 | 1421196_at 1427699_a_at 1451225_at | 12683 | 0.041 | -0.3040 | No | ||
| 47 | PLEKHA1 | 1417965_at 1443111_at | 12898 | 0.036 | -0.3135 | No | ||
| 48 | MAPK8 | 1420931_at 1420932_at 1437045_at 1440856_at 1457936_at | 13273 | 0.025 | -0.3304 | No | ||
| 49 | JUN | 1417409_at 1448694_at | 13314 | 0.024 | -0.3320 | No | ||
| 50 | BCAR1 | 1439388_s_at 1450622_at | 14577 | -0.017 | -0.3897 | No | ||
| 51 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 15150 | -0.039 | -0.4156 | No | ||
| 52 | GSK3A | 1435638_at | 16002 | -0.081 | -0.4538 | No | ||
| 53 | PTPN18 | 1419125_at 1444894_at | 16076 | -0.086 | -0.4564 | No | ||
| 54 | SHC1 | 1422853_at 1422854_at | 16122 | -0.088 | -0.4577 | No | ||
| 55 | REL | 1420710_at | 16411 | -0.106 | -0.4699 | No | ||
| 56 | GTF2I | 1431675_a_at 1431676_x_at | 16496 | -0.112 | -0.4727 | No | ||
| 57 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 16559 | -0.117 | -0.4745 | No | ||
| 58 | GAB1 | 1417693_a_at 1417694_at 1448814_at | 16627 | -0.123 | -0.4764 | No | ||
| 59 | HCK | 1446553_at 1449455_at | 16728 | -0.130 | -0.4798 | No | ||
| 60 | GRB2 | 1418508_a_at 1449111_a_at | 17171 | -0.170 | -0.4985 | No | ||
| 61 | CDK6 | 1435338_at 1440040_at 1455287_at 1460291_at | 17266 | -0.181 | -0.5012 | No | ||
| 62 | CHUK | 1417091_at 1428210_s_at 1451383_a_at | 17435 | -0.202 | -0.5071 | No | ||
| 63 | PRKCE | 1437860_at 1437861_s_at 1442609_at 1444649_at 1449956_at 1452878_at | 17444 | -0.203 | -0.5056 | No | ||
| 64 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 17487 | -0.209 | -0.5056 | No | ||
| 65 | ITPR2 | 1421678_at 1424833_at 1424834_s_at 1427287_s_at 1427693_at 1444418_at 1444551_at 1447328_at | 17601 | -0.226 | -0.5087 | No | ||
| 66 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 17944 | -0.270 | -0.5219 | No | ||
| 67 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 18001 | -0.278 | -0.5220 | No | ||
| 68 | CREB1 | 1421582_a_at 1421583_at 1423402_at 1428755_at 1452529_a_at 1452901_at | 18053 | -0.285 | -0.5217 | No | ||
| 69 | CDK2 | 1416873_a_at 1447617_at | 18435 | -0.345 | -0.5360 | No | ||
| 70 | CCNA2 | 1417910_at 1417911_at | 18439 | -0.346 | -0.5330 | No | ||
| 71 | SOS2 | 1452281_at | 18613 | -0.379 | -0.5375 | Yes | ||
| 72 | MAPK3 | 1427060_at | 18642 | -0.385 | -0.5352 | Yes | ||
| 73 | BCL2L11 | 1426334_a_at 1435448_at 1435449_at 1456005_a_at 1456006_at 1459794_at | 18691 | -0.394 | -0.5338 | Yes | ||
| 74 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 18818 | -0.421 | -0.5358 | Yes | ||
| 75 | NFATC2 | 1425901_at 1425990_a_at 1426031_a_at 1426032_at 1439205_at 1440426_at | 18829 | -0.424 | -0.5324 | Yes | ||
| 76 | DAPP1 | 1421936_at 1421937_at 1447337_at | 18854 | -0.431 | -0.5295 | Yes | ||
| 77 | HCLS1 | 1418842_at | 18958 | -0.455 | -0.5301 | Yes | ||
| 78 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 18974 | -0.459 | -0.5266 | Yes | ||
| 79 | SOS1 | 1421884_at 1421885_at 1421886_at | 18996 | -0.463 | -0.5234 | Yes | ||
| 80 | ACTR3 | 1426392_a_at 1434968_a_at 1452051_at | 19009 | -0.466 | -0.5197 | Yes | ||
| 81 | RAPGEF1 | 1421146_at 1427006_at 1441386_at | 19197 | -0.510 | -0.5236 | Yes | ||
| 82 | WAS | 1419631_at | 19333 | -0.543 | -0.5248 | Yes | ||
| 83 | HDAC5 | 1415743_at | 19351 | -0.547 | -0.5206 | Yes | ||
| 84 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 19447 | -0.577 | -0.5197 | Yes | ||
| 85 | ACTR2 | 1441660_at 1452587_at | 19498 | -0.591 | -0.5166 | Yes | ||
| 86 | LIME1 | 1416869_x_at 1437589_x_at 1448500_a_at | 19553 | -0.608 | -0.5135 | Yes | ||
| 87 | CASP9 | 1426125_a_at 1437537_at 1457816_at | 19786 | -0.679 | -0.5179 | Yes | ||
| 88 | RELA | 1419536_a_at | 20015 | -0.767 | -0.5214 | Yes | ||
| 89 | PTPN6 | 1456694_x_at 1460188_at | 20072 | -0.789 | -0.5168 | Yes | ||
| 90 | VAV1 | 1422932_a_at | 20423 | -0.958 | -0.5241 | Yes | ||
| 91 | IKBKG | 1421208_at 1421209_s_at 1435646_at 1435647_at 1450161_at 1454690_at | 20443 | -0.967 | -0.5161 | Yes | ||
| 92 | SLA2 | 1437504_at 1451492_at | 20516 | -1.005 | -0.5102 | Yes | ||
| 93 | PPP3CA | 1426401_at 1438478_a_at 1440051_at 1452056_s_at 1458495_at | 20582 | -1.046 | -0.5037 | Yes | ||
| 94 | ATF2 | 1426582_at 1426583_at 1427559_a_at 1437777_at 1452116_s_at 1459237_at | 20657 | -1.100 | -0.4970 | Yes | ||
| 95 | SH3BP2 | 1448328_at | 20677 | -1.112 | -0.4878 | Yes | ||
| 96 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 20696 | -1.127 | -0.4783 | Yes | ||
| 97 | CRKL | 1421953_at 1421954_at 1425604_at 1436950_at | 20749 | -1.171 | -0.4700 | Yes | ||
| 98 | GAB2 | 1419829_a_at 1420785_at 1439786_at 1446860_at | 20788 | -1.204 | -0.4608 | Yes | ||
| 99 | PDK2 | 1448825_at | 20844 | -1.246 | -0.4519 | Yes | ||
| 100 | PRKCQ | 1426044_a_at | 20851 | -1.253 | -0.4408 | Yes | ||
| 101 | CBLB | 1437304_at 1455082_at 1458469_at | 20915 | -1.305 | -0.4317 | Yes | ||
| 102 | CSK | 1423518_at 1439744_at | 21063 | -1.495 | -0.4248 | Yes | ||
| 103 | LCP2 | 1418641_at 1418642_at | 21182 | -1.674 | -0.4150 | Yes | ||
| 104 | CD79B | 1417640_at | 21236 | -1.761 | -0.4013 | Yes | ||
| 105 | CCND3 | 1415907_at 1437584_at 1444323_at 1446567_at 1447434_at 1457549_at | 21260 | -1.792 | -0.3860 | Yes | ||
| 106 | CARD11 | 1430257_at 1435996_at | 21299 | -1.844 | -0.3710 | Yes | ||
| 107 | NFATC3 | 1419976_s_at 1452497_a_at | 21381 | -1.965 | -0.3567 | Yes | ||
| 108 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 21477 | -2.135 | -0.3416 | Yes | ||
| 109 | NEDD9 | 1422818_at 1437132_x_at 1447885_x_at 1450767_at | 21487 | -2.153 | -0.3224 | Yes | ||
| 110 | ITK | 1417171_at 1430833_at 1452518_a_at 1456836_at 1457120_at | 21504 | -2.182 | -0.3032 | Yes | ||
| 111 | CD5 | 1418353_at | 21544 | -2.273 | -0.2843 | Yes | ||
| 112 | PRKCD | 1422847_a_at 1442256_at | 21582 | -2.392 | -0.2641 | Yes | ||
| 113 | CASP7 | 1426062_a_at 1448659_at | 21669 | -2.720 | -0.2433 | Yes | ||
| 114 | BCL6 | 1421818_at 1450381_a_at | 21730 | -2.913 | -0.2194 | Yes | ||
| 115 | PDPK1 | 1415729_at 1416501_at 1459775_at 1459776_x_at | 21761 | -3.077 | -0.1928 | Yes | ||
| 116 | PTK2 | 1423059_at 1430827_a_at 1439198_at 1440082_at 1441475_at 1443384_at 1445137_at | 21796 | -3.373 | -0.1635 | Yes | ||
| 117 | GSK3B | 1434439_at 1437001_at 1439931_at 1439949_at 1451020_at 1454958_at | 21811 | -3.432 | -0.1329 | Yes | ||
| 118 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21827 | -3.509 | -0.1015 | Yes | ||
| 119 | PTK2B | 1434653_at 1442437_at 1442927_at | 21836 | -3.576 | -0.0693 | Yes | ||
| 120 | VAV2 | 1421272_at 1435065_x_at 1435244_at 1458758_at | 21871 | -3.956 | -0.0347 | Yes | ||
| 121 | TEC | 1441524_at 1460204_at | 21881 | -4.130 | 0.0025 | Yes |