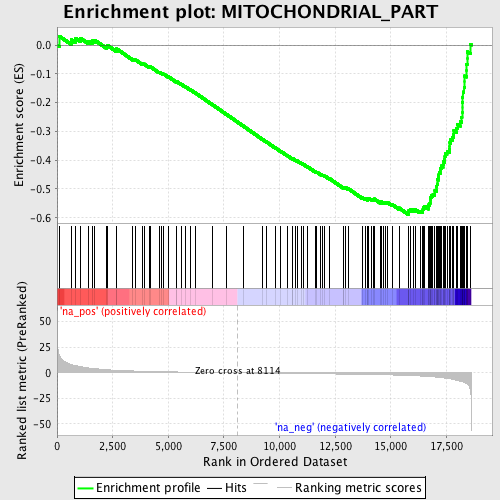
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MITOCHONDRIAL_PART |
| Enrichment Score (ES) | -0.58857334 |
| Normalized Enrichment Score (NES) | -2.1520889 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | RAB11FIP5 | 97 | 17.417 | 0.0311 | No | ||
| 2 | CASP7 | 643 | 8.075 | 0.0185 | No | ||
| 3 | COX6B2 | 819 | 7.059 | 0.0237 | No | ||
| 4 | SLC25A11 | 1051 | 6.015 | 0.0238 | No | ||
| 5 | SLC25A1 | 1418 | 4.797 | 0.0140 | No | ||
| 6 | ETFB | 1576 | 4.397 | 0.0147 | No | ||
| 7 | TIMM17B | 1687 | 4.111 | 0.0173 | No | ||
| 8 | MCL1 | 2240 | 3.055 | -0.0062 | No | ||
| 9 | NDUFS4 | 2245 | 3.040 | -0.0001 | No | ||
| 10 | UQCRH | 2674 | 2.461 | -0.0181 | No | ||
| 11 | NDUFA13 | 2679 | 2.452 | -0.0132 | No | ||
| 12 | MFN2 | 3405 | 1.812 | -0.0486 | No | ||
| 13 | HSD3B2 | 3526 | 1.725 | -0.0515 | No | ||
| 14 | FIS1 | 3826 | 1.540 | -0.0645 | No | ||
| 15 | NR3C1 | 3907 | 1.496 | -0.0657 | No | ||
| 16 | PITRM1 | 4144 | 1.357 | -0.0756 | No | ||
| 17 | CENTA2 | 4190 | 1.339 | -0.0753 | No | ||
| 18 | NDUFA1 | 4590 | 1.148 | -0.0945 | No | ||
| 19 | SDHA | 4695 | 1.103 | -0.0978 | No | ||
| 20 | CASQ1 | 4786 | 1.066 | -0.1004 | No | ||
| 21 | ALAS2 | 4990 | 0.981 | -0.1094 | No | ||
| 22 | UQCRB | 5353 | 0.847 | -0.1272 | No | ||
| 23 | VDAC1 | 5378 | 0.837 | -0.1267 | No | ||
| 24 | NDUFA2 | 5588 | 0.750 | -0.1365 | No | ||
| 25 | HCCS | 5753 | 0.697 | -0.1439 | No | ||
| 26 | MAOB | 5995 | 0.623 | -0.1556 | No | ||
| 27 | NDUFA6 | 6208 | 0.559 | -0.1659 | No | ||
| 28 | MRPS36 | 6964 | 0.334 | -0.2061 | No | ||
| 29 | NDUFS8 | 7597 | 0.146 | -0.2399 | No | ||
| 30 | GATM | 8358 | -0.072 | -0.2809 | No | ||
| 31 | BCKDHA | 9245 | -0.287 | -0.3282 | No | ||
| 32 | MRPS16 | 9418 | -0.331 | -0.3368 | No | ||
| 33 | ABCB7 | 9814 | -0.427 | -0.3573 | No | ||
| 34 | MTX2 | 10046 | -0.490 | -0.3688 | No | ||
| 35 | RHOT1 | 10355 | -0.570 | -0.3842 | No | ||
| 36 | PHB | 10585 | -0.623 | -0.3953 | No | ||
| 37 | PPOX | 10599 | -0.627 | -0.3947 | No | ||
| 38 | NNT | 10722 | -0.652 | -0.4000 | No | ||
| 39 | RAF1 | 10793 | -0.671 | -0.4024 | No | ||
| 40 | MRPL32 | 10973 | -0.710 | -0.4106 | No | ||
| 41 | MRPS22 | 11060 | -0.730 | -0.4137 | No | ||
| 42 | GRPEL1 | 11263 | -0.778 | -0.4230 | No | ||
| 43 | OGDH | 11610 | -0.871 | -0.4399 | No | ||
| 44 | ATP5G2 | 11669 | -0.882 | -0.4412 | No | ||
| 45 | DBT | 11859 | -0.932 | -0.4495 | No | ||
| 46 | ATP5C1 | 11946 | -0.956 | -0.4521 | No | ||
| 47 | ATP5J | 12033 | -0.980 | -0.4547 | No | ||
| 48 | NDUFS1 | 12247 | -1.037 | -0.4641 | No | ||
| 49 | MRPS21 | 12877 | -1.223 | -0.4956 | No | ||
| 50 | MRPS18C | 12885 | -1.226 | -0.4934 | No | ||
| 51 | MPV17 | 12944 | -1.243 | -0.4939 | No | ||
| 52 | RHOT2 | 13078 | -1.284 | -0.4984 | No | ||
| 53 | SLC25A3 | 13705 | -1.491 | -0.5292 | No | ||
| 54 | BNIP3 | 13848 | -1.550 | -0.5336 | No | ||
| 55 | ATP5E | 13959 | -1.590 | -0.5363 | No | ||
| 56 | BCKDK | 13967 | -1.593 | -0.5333 | No | ||
| 57 | UCP3 | 14018 | -1.610 | -0.5327 | No | ||
| 58 | UQCRC1 | 14129 | -1.654 | -0.5352 | No | ||
| 59 | MRPL51 | 14225 | -1.688 | -0.5368 | No | ||
| 60 | CS | 14249 | -1.697 | -0.5345 | No | ||
| 61 | VDAC3 | 14545 | -1.833 | -0.5466 | No | ||
| 62 | TIMM23 | 14576 | -1.845 | -0.5444 | No | ||
| 63 | HSD3B1 | 14677 | -1.891 | -0.5459 | No | ||
| 64 | ATP5B | 14762 | -1.928 | -0.5464 | No | ||
| 65 | SURF1 | 14828 | -1.958 | -0.5458 | No | ||
| 66 | TIMM9 | 15058 | -2.078 | -0.5539 | No | ||
| 67 | ATP5F1 | 15369 | -2.265 | -0.5659 | No | ||
| 68 | ATP5O | 15789 | -2.572 | -0.5832 | Yes | ||
| 69 | AIFM2 | 15796 | -2.585 | -0.5781 | Yes | ||
| 70 | ATP5G3 | 15811 | -2.601 | -0.5735 | Yes | ||
| 71 | COX15 | 15890 | -2.670 | -0.5721 | Yes | ||
| 72 | SLC25A22 | 15997 | -2.778 | -0.5721 | Yes | ||
| 73 | PIN4 | 16097 | -2.867 | -0.5714 | Yes | ||
| 74 | MRPL52 | 16313 | -3.080 | -0.5766 | Yes | ||
| 75 | ATP5A1 | 16439 | -3.220 | -0.5767 | Yes | ||
| 76 | TOMM34 | 16443 | -3.224 | -0.5701 | Yes | ||
| 77 | ACN9 | 16474 | -3.276 | -0.5649 | Yes | ||
| 78 | NDUFS2 | 16515 | -3.325 | -0.5601 | Yes | ||
| 79 | ATP5D | 16693 | -3.589 | -0.5622 | Yes | ||
| 80 | OPA1 | 16708 | -3.612 | -0.5554 | Yes | ||
| 81 | NDUFAB1 | 16750 | -3.672 | -0.5500 | Yes | ||
| 82 | MRPS11 | 16765 | -3.711 | -0.5430 | Yes | ||
| 83 | BCL2 | 16768 | -3.720 | -0.5354 | Yes | ||
| 84 | MRPL55 | 16796 | -3.760 | -0.5290 | Yes | ||
| 85 | NDUFS7 | 16839 | -3.824 | -0.5233 | Yes | ||
| 86 | ABCB6 | 16893 | -3.909 | -0.5180 | Yes | ||
| 87 | BCKDHB | 16980 | -4.054 | -0.5142 | Yes | ||
| 88 | DNAJA3 | 16981 | -4.055 | -0.5057 | Yes | ||
| 89 | TIMM8B | 17052 | -4.213 | -0.5007 | Yes | ||
| 90 | HADHB | 17057 | -4.223 | -0.4921 | Yes | ||
| 91 | ETFA | 17075 | -4.248 | -0.4842 | Yes | ||
| 92 | TFB2M | 17101 | -4.311 | -0.4766 | Yes | ||
| 93 | ACADM | 17102 | -4.311 | -0.4676 | Yes | ||
| 94 | SDHD | 17130 | -4.383 | -0.4599 | Yes | ||
| 95 | TOMM22 | 17141 | -4.400 | -0.4512 | Yes | ||
| 96 | NDUFA9 | 17165 | -4.464 | -0.4432 | Yes | ||
| 97 | HTRA2 | 17219 | -4.583 | -0.4365 | Yes | ||
| 98 | NDUFV1 | 17230 | -4.610 | -0.4274 | Yes | ||
| 99 | MRPS28 | 17273 | -4.704 | -0.4199 | Yes | ||
| 100 | ALDH4A1 | 17347 | -4.890 | -0.4136 | Yes | ||
| 101 | TIMM50 | 17381 | -4.986 | -0.4050 | Yes | ||
| 102 | TIMM44 | 17416 | -5.062 | -0.3963 | Yes | ||
| 103 | MRPS15 | 17432 | -5.110 | -0.3864 | Yes | ||
| 104 | PMPCA | 17453 | -5.184 | -0.3767 | Yes | ||
| 105 | MRPL23 | 17526 | -5.410 | -0.3693 | Yes | ||
| 106 | MRPL40 | 17631 | -5.748 | -0.3629 | Yes | ||
| 107 | ATP5G1 | 17650 | -5.797 | -0.3518 | Yes | ||
| 108 | MRPS18A | 17654 | -5.807 | -0.3399 | Yes | ||
| 109 | MRPS12 | 17691 | -5.951 | -0.3294 | Yes | ||
| 110 | VDAC2 | 17759 | -6.247 | -0.3200 | Yes | ||
| 111 | BCS1L | 17809 | -6.491 | -0.3091 | Yes | ||
| 112 | MRPL10 | 17834 | -6.574 | -0.2967 | Yes | ||
| 113 | MRPS35 | 17954 | -7.161 | -0.2882 | Yes | ||
| 114 | IMMT | 17995 | -7.360 | -0.2750 | Yes | ||
| 115 | POLG2 | 18115 | -8.011 | -0.2647 | Yes | ||
| 116 | CYCS | 18181 | -8.429 | -0.2507 | Yes | ||
| 117 | SLC25A15 | 18200 | -8.547 | -0.2338 | Yes | ||
| 118 | SUPV3L1 | 18207 | -8.593 | -0.2162 | Yes | ||
| 119 | NDUFS3 | 18208 | -8.602 | -0.1983 | Yes | ||
| 120 | TIMM17A | 18233 | -8.852 | -0.1811 | Yes | ||
| 121 | TIMM10 | 18244 | -8.958 | -0.1629 | Yes | ||
| 122 | BAX | 18289 | -9.342 | -0.1458 | Yes | ||
| 123 | MRPS24 | 18323 | -9.661 | -0.1275 | Yes | ||
| 124 | NFS1 | 18325 | -9.674 | -0.1073 | Yes | ||
| 125 | PHB2 | 18395 | -10.466 | -0.0892 | Yes | ||
| 126 | ABCF2 | 18400 | -10.546 | -0.0675 | Yes | ||
| 127 | MRPL12 | 18432 | -10.904 | -0.0464 | Yes | ||
| 128 | MRPS10 | 18436 | -10.998 | -0.0236 | Yes | ||
| 129 | TIMM13 | 18569 | -15.957 | 0.0025 | Yes |
