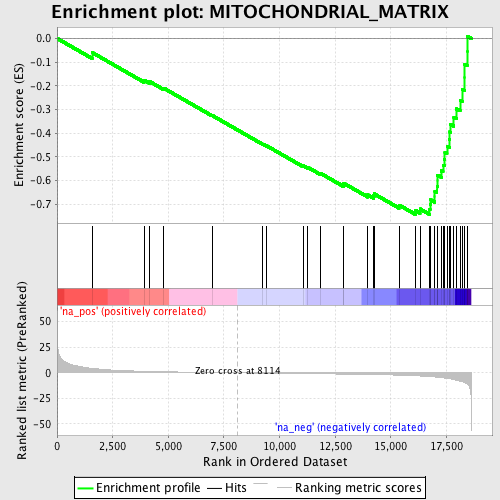
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | MITOCHONDRIAL_MATRIX |
| Enrichment Score (ES) | -0.7433782 |
| Normalized Enrichment Score (NES) | -2.250874 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.0 |
| FWER p-Value | 0.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ETFB | 1576 | 4.397 | -0.0595 | No | ||
| 2 | NR3C1 | 3907 | 1.496 | -0.1764 | No | ||
| 3 | PITRM1 | 4144 | 1.357 | -0.1813 | No | ||
| 4 | CASQ1 | 4786 | 1.066 | -0.2096 | No | ||
| 5 | MRPS36 | 6964 | 0.334 | -0.3249 | No | ||
| 6 | BCKDHA | 9245 | -0.287 | -0.4460 | No | ||
| 7 | MRPS16 | 9418 | -0.331 | -0.4534 | No | ||
| 8 | MRPS22 | 11060 | -0.730 | -0.5375 | No | ||
| 9 | GRPEL1 | 11263 | -0.778 | -0.5439 | No | ||
| 10 | DBT | 11859 | -0.932 | -0.5706 | No | ||
| 11 | MRPS21 | 12877 | -1.223 | -0.6183 | No | ||
| 12 | MRPS18C | 12885 | -1.226 | -0.6116 | No | ||
| 13 | BCKDK | 13967 | -1.593 | -0.6606 | No | ||
| 14 | MRPL51 | 14225 | -1.688 | -0.6647 | No | ||
| 15 | CS | 14249 | -1.697 | -0.6562 | No | ||
| 16 | ATP5F1 | 15369 | -2.265 | -0.7034 | No | ||
| 17 | PIN4 | 16097 | -2.867 | -0.7261 | Yes | ||
| 18 | MRPL52 | 16313 | -3.080 | -0.7199 | Yes | ||
| 19 | NDUFAB1 | 16750 | -3.672 | -0.7222 | Yes | ||
| 20 | MRPS11 | 16765 | -3.711 | -0.7016 | Yes | ||
| 21 | MRPL55 | 16796 | -3.760 | -0.6816 | Yes | ||
| 22 | BCKDHB | 16980 | -4.054 | -0.6681 | Yes | ||
| 23 | DNAJA3 | 16981 | -4.055 | -0.6448 | Yes | ||
| 24 | ETFA | 17075 | -4.248 | -0.6253 | Yes | ||
| 25 | TFB2M | 17101 | -4.311 | -0.6019 | Yes | ||
| 26 | ACADM | 17102 | -4.311 | -0.5771 | Yes | ||
| 27 | MRPS28 | 17273 | -4.704 | -0.5591 | Yes | ||
| 28 | ALDH4A1 | 17347 | -4.890 | -0.5349 | Yes | ||
| 29 | TIMM44 | 17416 | -5.062 | -0.5094 | Yes | ||
| 30 | MRPS15 | 17432 | -5.110 | -0.4808 | Yes | ||
| 31 | MRPL23 | 17526 | -5.410 | -0.4547 | Yes | ||
| 32 | MRPL40 | 17631 | -5.748 | -0.4272 | Yes | ||
| 33 | MRPS18A | 17654 | -5.807 | -0.3950 | Yes | ||
| 34 | MRPS12 | 17691 | -5.951 | -0.3626 | Yes | ||
| 35 | MRPL10 | 17834 | -6.574 | -0.3324 | Yes | ||
| 36 | MRPS35 | 17954 | -7.161 | -0.2976 | Yes | ||
| 37 | POLG2 | 18115 | -8.011 | -0.2601 | Yes | ||
| 38 | SUPV3L1 | 18207 | -8.593 | -0.2156 | Yes | ||
| 39 | MRPS24 | 18323 | -9.661 | -0.1661 | Yes | ||
| 40 | NFS1 | 18325 | -9.674 | -0.1105 | Yes | ||
| 41 | MRPL12 | 18432 | -10.904 | -0.0535 | Yes | ||
| 42 | MRPS10 | 18436 | -10.998 | 0.0097 | Yes |
