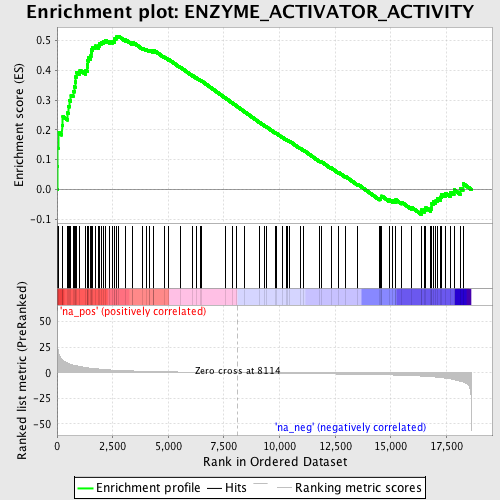
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | ENZYME_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | 0.5145407 |
| Normalized Enrichment Score (NES) | 1.6868421 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.22824743 |
| FWER p-Value | 0.989 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 12 | 30.494 | 0.0762 | Yes | ||
| 2 | ABR | 25 | 25.496 | 0.1399 | Yes | ||
| 3 | PRKRA | 47 | 21.083 | 0.1919 | Yes | ||
| 4 | GMFG | 219 | 13.087 | 0.2156 | Yes | ||
| 5 | THY1 | 260 | 12.306 | 0.2445 | Yes | ||
| 6 | ARFGAP3 | 476 | 9.537 | 0.2569 | Yes | ||
| 7 | BCR | 501 | 9.330 | 0.2792 | Yes | ||
| 8 | RGS12 | 549 | 8.811 | 0.2988 | Yes | ||
| 9 | ALOX5AP | 623 | 8.201 | 0.3156 | Yes | ||
| 10 | RASA4 | 720 | 7.607 | 0.3296 | Yes | ||
| 11 | NOD1 | 769 | 7.328 | 0.3454 | Yes | ||
| 12 | SIPA1 | 813 | 7.080 | 0.3610 | Yes | ||
| 13 | RALBP1 | 832 | 7.008 | 0.3777 | Yes | ||
| 14 | GUCA1A | 856 | 6.933 | 0.3939 | Yes | ||
| 15 | RASA3 | 1025 | 6.143 | 0.4003 | Yes | ||
| 16 | PSAP | 1291 | 5.146 | 0.3990 | Yes | ||
| 17 | MAP3K5 | 1357 | 4.952 | 0.4079 | Yes | ||
| 18 | RGS3 | 1364 | 4.932 | 0.4200 | Yes | ||
| 19 | CDC42EP2 | 1379 | 4.896 | 0.4316 | Yes | ||
| 20 | ARHGAP10 | 1417 | 4.798 | 0.4417 | Yes | ||
| 21 | CASP1 | 1513 | 4.546 | 0.4481 | Yes | ||
| 22 | FZR1 | 1528 | 4.521 | 0.4587 | Yes | ||
| 23 | RGS2 | 1552 | 4.449 | 0.4687 | Yes | ||
| 24 | RABGAP1 | 1596 | 4.330 | 0.4773 | Yes | ||
| 25 | RASA1 | 1707 | 4.056 | 0.4815 | Yes | ||
| 26 | PRKCD | 1875 | 3.701 | 0.4819 | Yes | ||
| 27 | OPHN1 | 1892 | 3.648 | 0.4902 | Yes | ||
| 28 | ALS2 | 1980 | 3.491 | 0.4943 | Yes | ||
| 29 | RGS1 | 2085 | 3.285 | 0.4970 | Yes | ||
| 30 | ARHGAP4 | 2163 | 3.163 | 0.5008 | Yes | ||
| 31 | GHRL | 2348 | 2.851 | 0.4980 | Yes | ||
| 32 | FGF13 | 2478 | 2.679 | 0.4978 | Yes | ||
| 33 | MYO9B | 2569 | 2.584 | 0.4995 | Yes | ||
| 34 | RGS9 | 2583 | 2.568 | 0.5052 | Yes | ||
| 35 | CHN2 | 2649 | 2.487 | 0.5080 | Yes | ||
| 36 | IGFBP3 | 2685 | 2.446 | 0.5123 | Yes | ||
| 37 | GUCA1C | 2755 | 2.379 | 0.5145 | Yes | ||
| 38 | HSPB2 | 3071 | 2.078 | 0.5028 | No | ||
| 39 | DNMT3L | 3382 | 1.826 | 0.4906 | No | ||
| 40 | BCL10 | 3391 | 1.822 | 0.4948 | No | ||
| 41 | F10 | 3844 | 1.530 | 0.4742 | No | ||
| 42 | NF1 | 4013 | 1.433 | 0.4688 | No | ||
| 43 | PITRM1 | 4144 | 1.357 | 0.4652 | No | ||
| 44 | IL2 | 4161 | 1.353 | 0.4677 | No | ||
| 45 | RGS6 | 4324 | 1.264 | 0.4622 | No | ||
| 46 | RCVRN | 4331 | 1.260 | 0.4650 | No | ||
| 47 | MADD | 4336 | 1.256 | 0.4680 | No | ||
| 48 | APOA5 | 4820 | 1.054 | 0.4445 | No | ||
| 49 | ARL1 | 5018 | 0.973 | 0.4363 | No | ||
| 50 | CASP8AP2 | 5544 | 0.772 | 0.4099 | No | ||
| 51 | SOS1 | 6098 | 0.595 | 0.3816 | No | ||
| 52 | RACGAP1 | 6256 | 0.543 | 0.3745 | No | ||
| 53 | DOCK4 | 6432 | 0.492 | 0.3662 | No | ||
| 54 | APOA4 | 6498 | 0.469 | 0.3639 | No | ||
| 55 | ARFGEF1 | 7590 | 0.148 | 0.3054 | No | ||
| 56 | TSC2 | 7895 | 0.062 | 0.2891 | No | ||
| 57 | DNAJC1 | 8047 | 0.016 | 0.2810 | No | ||
| 58 | RGS11 | 8441 | -0.091 | 0.2600 | No | ||
| 59 | RASGRP3 | 9100 | -0.251 | 0.2251 | No | ||
| 60 | MMP15 | 9330 | -0.309 | 0.2135 | No | ||
| 61 | RGS20 | 9403 | -0.327 | 0.2104 | No | ||
| 62 | GMFB | 9805 | -0.425 | 0.1898 | No | ||
| 63 | MMP24 | 9867 | -0.441 | 0.1877 | No | ||
| 64 | ALDH1A1 | 10135 | -0.511 | 0.1745 | No | ||
| 65 | TPD52L1 | 10295 | -0.556 | 0.1673 | No | ||
| 66 | ARHGAP5 | 10358 | -0.571 | 0.1654 | No | ||
| 67 | RASA2 | 10439 | -0.590 | 0.1626 | No | ||
| 68 | GUCA1B | 10930 | -0.702 | 0.1379 | No | ||
| 69 | MMP16 | 11074 | -0.733 | 0.1320 | No | ||
| 70 | DEPDC2 | 11797 | -0.915 | 0.0953 | No | ||
| 71 | TRIM23 | 11893 | -0.942 | 0.0926 | No | ||
| 72 | APOC2 | 12310 | -1.052 | 0.0728 | No | ||
| 73 | GUCA2B | 12664 | -1.161 | 0.0566 | No | ||
| 74 | RGS4 | 12983 | -1.257 | 0.0426 | No | ||
| 75 | RGS16 | 13520 | -1.431 | 0.0173 | No | ||
| 76 | CENTD2 | 14496 | -1.810 | -0.0308 | No | ||
| 77 | RFC1 | 14556 | -1.839 | -0.0294 | No | ||
| 78 | PPP2R4 | 14558 | -1.840 | -0.0248 | No | ||
| 79 | CXCL1 | 14592 | -1.851 | -0.0219 | No | ||
| 80 | DLC1 | 14919 | -2.008 | -0.0344 | No | ||
| 81 | PLAA | 15076 | -2.085 | -0.0376 | No | ||
| 82 | RANGAP1 | 15217 | -2.168 | -0.0397 | No | ||
| 83 | APOA2 | 15226 | -2.172 | -0.0347 | No | ||
| 84 | MAPK8IP2 | 15488 | -2.341 | -0.0429 | No | ||
| 85 | MMP17 | 15922 | -2.707 | -0.0594 | No | ||
| 86 | RASAL1 | 16377 | -3.139 | -0.0760 | No | ||
| 87 | CTSA | 16386 | -3.149 | -0.0685 | No | ||
| 88 | DNAJB6 | 16525 | -3.337 | -0.0676 | No | ||
| 89 | APOA1 | 16575 | -3.406 | -0.0616 | No | ||
| 90 | DBNL | 16799 | -3.764 | -0.0642 | No | ||
| 91 | GMIP | 16837 | -3.820 | -0.0566 | No | ||
| 92 | RGS14 | 16840 | -3.824 | -0.0470 | No | ||
| 93 | BNIP2 | 16914 | -3.943 | -0.0410 | No | ||
| 94 | CENTA1 | 17027 | -4.161 | -0.0366 | No | ||
| 95 | ADRM1 | 17092 | -4.293 | -0.0292 | No | ||
| 96 | CASP9 | 17236 | -4.631 | -0.0253 | No | ||
| 97 | CD24 | 17278 | -4.710 | -0.0156 | No | ||
| 98 | GTF3C4 | 17454 | -5.193 | -0.0120 | No | ||
| 99 | MALT1 | 17693 | -5.961 | -0.0098 | No | ||
| 100 | AHSA1 | 17842 | -6.603 | -0.0011 | No | ||
| 101 | CTAGE5 | 18119 | -8.020 | 0.0042 | No | ||
| 102 | CDK5R1 | 18246 | -8.970 | 0.0200 | No |