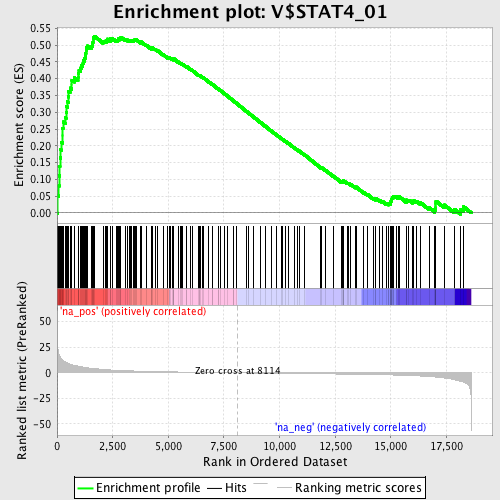
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$STAT4_01 |
| Enrichment Score (ES) | 0.52673006 |
| Normalized Enrichment Score (NES) | 1.8458447 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.025430147 |
| FWER p-Value | 0.071 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | VAV3 | 12 | 30.494 | 0.0524 | Yes | ||
| 2 | CA3 | 82 | 18.400 | 0.0807 | Yes | ||
| 3 | PRKCH | 92 | 17.715 | 0.1110 | Yes | ||
| 4 | SGK | 116 | 16.366 | 0.1383 | Yes | ||
| 5 | EDG5 | 131 | 15.570 | 0.1646 | Yes | ||
| 6 | CISH | 161 | 14.580 | 0.1884 | Yes | ||
| 7 | SATB1 | 187 | 13.719 | 0.2109 | Yes | ||
| 8 | BCL6B | 243 | 12.592 | 0.2298 | Yes | ||
| 9 | BLNK | 244 | 12.553 | 0.2517 | Yes | ||
| 10 | MEF2C | 271 | 12.059 | 0.2712 | Yes | ||
| 11 | OPRM1 | 383 | 10.560 | 0.2836 | Yes | ||
| 12 | ERG | 405 | 10.291 | 0.3004 | Yes | ||
| 13 | LAT | 424 | 10.053 | 0.3169 | Yes | ||
| 14 | HHEX | 462 | 9.718 | 0.3318 | Yes | ||
| 15 | PDE6D | 500 | 9.336 | 0.3460 | Yes | ||
| 16 | PDE4D | 515 | 9.182 | 0.3612 | Yes | ||
| 17 | PPARGC1A | 584 | 8.495 | 0.3723 | Yes | ||
| 18 | PDLIM2 | 663 | 7.927 | 0.3819 | Yes | ||
| 19 | HOXB4 | 664 | 7.921 | 0.3957 | Yes | ||
| 20 | FEZ2 | 772 | 7.311 | 0.4026 | Yes | ||
| 21 | LEPROTL1 | 940 | 6.568 | 0.4050 | Yes | ||
| 22 | STAT5A | 947 | 6.564 | 0.4161 | Yes | ||
| 23 | GNB2 | 981 | 6.352 | 0.4253 | Yes | ||
| 24 | TNFRSF19 | 1056 | 6.003 | 0.4318 | Yes | ||
| 25 | MAP4K2 | 1094 | 5.837 | 0.4399 | Yes | ||
| 26 | ZDHHC8 | 1160 | 5.551 | 0.4461 | Yes | ||
| 27 | HSD11B1 | 1190 | 5.453 | 0.4540 | Yes | ||
| 28 | IGF1 | 1226 | 5.346 | 0.4614 | Yes | ||
| 29 | CASKIN2 | 1262 | 5.235 | 0.4686 | Yes | ||
| 30 | NOTCH4 | 1285 | 5.173 | 0.4764 | Yes | ||
| 31 | MBNL1 | 1300 | 5.110 | 0.4845 | Yes | ||
| 32 | RIN2 | 1310 | 5.083 | 0.4929 | Yes | ||
| 33 | PBX1 | 1383 | 4.889 | 0.4975 | Yes | ||
| 34 | GPHN | 1543 | 4.476 | 0.4967 | Yes | ||
| 35 | SDCBP | 1586 | 4.357 | 0.5020 | Yes | ||
| 36 | ARRDC3 | 1606 | 4.295 | 0.5084 | Yes | ||
| 37 | TCF15 | 1619 | 4.260 | 0.5152 | Yes | ||
| 38 | NFAT5 | 1621 | 4.255 | 0.5225 | Yes | ||
| 39 | TRPC4AP | 1677 | 4.127 | 0.5267 | Yes | ||
| 40 | MPO | 2064 | 3.325 | 0.5116 | No | ||
| 41 | CSRP3 | 2155 | 3.173 | 0.5122 | No | ||
| 42 | PRKCB1 | 2223 | 3.074 | 0.5139 | No | ||
| 43 | LIMK2 | 2244 | 3.043 | 0.5182 | No | ||
| 44 | LPL | 2393 | 2.780 | 0.5150 | No | ||
| 45 | USP2 | 2397 | 2.776 | 0.5196 | No | ||
| 46 | RBMS1 | 2497 | 2.662 | 0.5189 | No | ||
| 47 | DHH | 2653 | 2.483 | 0.5148 | No | ||
| 48 | SMOX | 2730 | 2.403 | 0.5149 | No | ||
| 49 | FEM1C | 2738 | 2.395 | 0.5187 | No | ||
| 50 | CPA3 | 2806 | 2.322 | 0.5191 | No | ||
| 51 | TNKS1BP1 | 2842 | 2.285 | 0.5212 | No | ||
| 52 | EGF | 2866 | 2.266 | 0.5239 | No | ||
| 53 | SLC38A2 | 3058 | 2.089 | 0.5171 | No | ||
| 54 | PTK7 | 3155 | 2.010 | 0.5154 | No | ||
| 55 | TBC1D17 | 3241 | 1.944 | 0.5142 | No | ||
| 56 | ITGA8 | 3289 | 1.909 | 0.5150 | No | ||
| 57 | PITPNC1 | 3359 | 1.846 | 0.5145 | No | ||
| 58 | FN1 | 3426 | 1.799 | 0.5140 | No | ||
| 59 | LNX2 | 3458 | 1.780 | 0.5154 | No | ||
| 60 | FLT1 | 3503 | 1.740 | 0.5161 | No | ||
| 61 | MRPL24 | 3553 | 1.710 | 0.5164 | No | ||
| 62 | PAX8 | 3752 | 1.590 | 0.5084 | No | ||
| 63 | PHF15 | 3782 | 1.567 | 0.5096 | No | ||
| 64 | STAB1 | 4003 | 1.435 | 0.5001 | No | ||
| 65 | PITX2 | 4241 | 1.312 | 0.4896 | No | ||
| 66 | RUNX2 | 4264 | 1.301 | 0.4906 | No | ||
| 67 | HOXD8 | 4268 | 1.298 | 0.4927 | No | ||
| 68 | MITF | 4405 | 1.227 | 0.4875 | No | ||
| 69 | PKP4 | 4523 | 1.172 | 0.4832 | No | ||
| 70 | FANK1 | 4761 | 1.079 | 0.4722 | No | ||
| 71 | VIT | 4980 | 0.986 | 0.4621 | No | ||
| 72 | MYL3 | 5034 | 0.967 | 0.4609 | No | ||
| 73 | CITED2 | 5036 | 0.967 | 0.4626 | No | ||
| 74 | HOXC6 | 5103 | 0.941 | 0.4606 | No | ||
| 75 | PHTF1 | 5182 | 0.913 | 0.4580 | No | ||
| 76 | UBE2D3 | 5234 | 0.895 | 0.4568 | No | ||
| 77 | NDFIP1 | 5237 | 0.894 | 0.4582 | No | ||
| 78 | CACNA1E | 5252 | 0.885 | 0.4590 | No | ||
| 79 | FEV | 5446 | 0.814 | 0.4500 | No | ||
| 80 | HNF4G | 5550 | 0.769 | 0.4457 | No | ||
| 81 | GIF | 5612 | 0.741 | 0.4437 | No | ||
| 82 | BMP5 | 5649 | 0.731 | 0.4430 | No | ||
| 83 | SESTD1 | 5815 | 0.677 | 0.4352 | No | ||
| 84 | FGF12 | 5820 | 0.676 | 0.4362 | No | ||
| 85 | JMJD1C | 6004 | 0.618 | 0.4274 | No | ||
| 86 | NEXN | 6083 | 0.598 | 0.4242 | No | ||
| 87 | CRYGB | 6347 | 0.515 | 0.4108 | No | ||
| 88 | RHOJ | 6378 | 0.508 | 0.4101 | No | ||
| 89 | HNRPD | 6399 | 0.500 | 0.4099 | No | ||
| 90 | DOCK4 | 6432 | 0.492 | 0.4090 | No | ||
| 91 | HIPK3 | 6531 | 0.459 | 0.4045 | No | ||
| 92 | CD3D | 6591 | 0.444 | 0.4020 | No | ||
| 93 | NCAM1 | 6819 | 0.376 | 0.3904 | No | ||
| 94 | HOXD10 | 6977 | 0.329 | 0.3824 | No | ||
| 95 | GREM1 | 6978 | 0.328 | 0.3830 | No | ||
| 96 | EPM2A | 7236 | 0.259 | 0.3695 | No | ||
| 97 | CALR | 7256 | 0.252 | 0.3689 | No | ||
| 98 | GSC | 7334 | 0.228 | 0.3652 | No | ||
| 99 | ARPC2 | 7517 | 0.170 | 0.3556 | No | ||
| 100 | STMN2 | 7648 | 0.132 | 0.3488 | No | ||
| 101 | PIK3C2G | 7936 | 0.050 | 0.3333 | No | ||
| 102 | CTSK | 7948 | 0.047 | 0.3328 | No | ||
| 103 | VSNL1 | 8042 | 0.018 | 0.3278 | No | ||
| 104 | NR4A1 | 8495 | -0.105 | 0.3035 | No | ||
| 105 | CACNG3 | 8624 | -0.136 | 0.2968 | No | ||
| 106 | FLI1 | 8812 | -0.183 | 0.2869 | No | ||
| 107 | HOXC13 | 9157 | -0.267 | 0.2688 | No | ||
| 108 | MAP2K7 | 9344 | -0.312 | 0.2592 | No | ||
| 109 | SLC26A3 | 9625 | -0.377 | 0.2447 | No | ||
| 110 | ERF | 9871 | -0.443 | 0.2322 | No | ||
| 111 | GRID2 | 10073 | -0.497 | 0.2221 | No | ||
| 112 | CSPG4 | 10134 | -0.511 | 0.2198 | No | ||
| 113 | PHOX2B | 10261 | -0.547 | 0.2139 | No | ||
| 114 | ETS1 | 10418 | -0.585 | 0.2065 | No | ||
| 115 | IL21 | 10658 | -0.640 | 0.1946 | No | ||
| 116 | LGI1 | 10823 | -0.678 | 0.1869 | No | ||
| 117 | MAPK10 | 10886 | -0.694 | 0.1848 | No | ||
| 118 | GPRC5D | 11116 | -0.743 | 0.1736 | No | ||
| 119 | PRCC | 11858 | -0.932 | 0.1351 | No | ||
| 120 | DRD1IP | 11867 | -0.935 | 0.1363 | No | ||
| 121 | NUDT4 | 12060 | -0.987 | 0.1276 | No | ||
| 122 | EVI1 | 12416 | -1.086 | 0.1102 | No | ||
| 123 | SLMAP | 12771 | -1.190 | 0.0931 | No | ||
| 124 | BDNF | 12829 | -1.208 | 0.0921 | No | ||
| 125 | TDRD9 | 12850 | -1.214 | 0.0932 | No | ||
| 126 | GRIN2B | 12859 | -1.218 | 0.0948 | No | ||
| 127 | POU4F1 | 12866 | -1.221 | 0.0966 | No | ||
| 128 | YARS | 13057 | -1.279 | 0.0886 | No | ||
| 129 | MTUS1 | 13089 | -1.287 | 0.0891 | No | ||
| 130 | NRG1 | 13205 | -1.321 | 0.0852 | No | ||
| 131 | RHOQ | 13408 | -1.392 | 0.0767 | No | ||
| 132 | ANGPTL1 | 13437 | -1.401 | 0.0776 | No | ||
| 133 | FGF10 | 13782 | -1.517 | 0.0616 | No | ||
| 134 | HSPB7 | 13957 | -1.589 | 0.0549 | No | ||
| 135 | TOM1L1 | 14216 | -1.684 | 0.0438 | No | ||
| 136 | HOXB9 | 14320 | -1.722 | 0.0413 | No | ||
| 137 | CSF3R | 14327 | -1.731 | 0.0439 | No | ||
| 138 | RUNX3 | 14506 | -1.813 | 0.0374 | No | ||
| 139 | SLC36A2 | 14644 | -1.874 | 0.0333 | No | ||
| 140 | TLL1 | 14812 | -1.953 | 0.0276 | No | ||
| 141 | HTR3B | 14886 | -1.992 | 0.0271 | No | ||
| 142 | A2BP1 | 14906 | -2.002 | 0.0296 | No | ||
| 143 | FGF8 | 14976 | -2.035 | 0.0294 | No | ||
| 144 | GRIA1 | 14984 | -2.039 | 0.0326 | No | ||
| 145 | TRERF1 | 14999 | -2.046 | 0.0354 | No | ||
| 146 | BZW1 | 15016 | -2.054 | 0.0381 | No | ||
| 147 | TMC3 | 15036 | -2.067 | 0.0406 | No | ||
| 148 | RNF138 | 15051 | -2.075 | 0.0435 | No | ||
| 149 | SHOX2 | 15065 | -2.080 | 0.0464 | No | ||
| 150 | UBL3 | 15100 | -2.103 | 0.0482 | No | ||
| 151 | CMYA5 | 15141 | -2.129 | 0.0498 | No | ||
| 152 | EML4 | 15235 | -2.176 | 0.0485 | No | ||
| 153 | HOXB3 | 15359 | -2.260 | 0.0458 | No | ||
| 154 | AKT1S1 | 15371 | -2.265 | 0.0491 | No | ||
| 155 | POU3F3 | 15720 | -2.510 | 0.0346 | No | ||
| 156 | GPX1 | 15722 | -2.510 | 0.0389 | No | ||
| 157 | AP1G2 | 15813 | -2.603 | 0.0386 | No | ||
| 158 | EDN1 | 15985 | -2.766 | 0.0341 | No | ||
| 159 | OSR1 | 16036 | -2.815 | 0.0363 | No | ||
| 160 | ACBD6 | 16153 | -2.927 | 0.0351 | No | ||
| 161 | SOSTDC1 | 16326 | -3.096 | 0.0312 | No | ||
| 162 | POLR1D | 16717 | -3.627 | 0.0163 | No | ||
| 163 | TTYH2 | 16978 | -4.052 | 0.0093 | No | ||
| 164 | NEDD4 | 17006 | -4.114 | 0.0150 | No | ||
| 165 | TWIST1 | 17022 | -4.143 | 0.0214 | No | ||
| 166 | NRAS | 17023 | -4.143 | 0.0286 | No | ||
| 167 | CENTA1 | 17027 | -4.161 | 0.0357 | No | ||
| 168 | CLOCK | 17406 | -5.039 | 0.0239 | No | ||
| 169 | LRRC16 | 17849 | -6.624 | 0.0115 | No | ||
| 170 | OSGEPL1 | 18145 | -8.182 | 0.0098 | No | ||
| 171 | EXTL2 | 18255 | -9.040 | 0.0196 | No |
