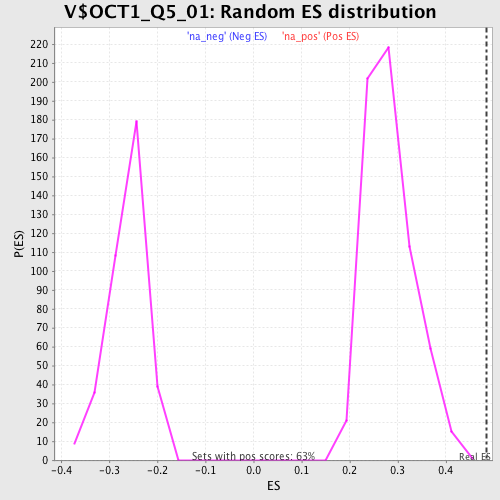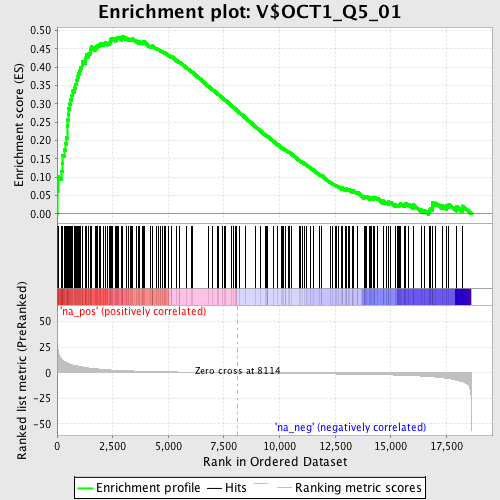
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$OCT1_Q5_01 |
| Enrichment Score (ES) | 0.48481873 |
| Normalized Enrichment Score (NES) | 1.7190686 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.054183103 |
| FWER p-Value | 0.365 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | STAT4 | 6 | 35.873 | 0.0637 | Yes | ||
| 2 | HOXA7 | 40 | 22.177 | 0.1015 | Yes | ||
| 3 | VPREB3 | 210 | 13.301 | 0.1161 | Yes | ||
| 4 | GPRC5B | 234 | 12.715 | 0.1376 | Yes | ||
| 5 | BLNK | 244 | 12.553 | 0.1595 | Yes | ||
| 6 | CYR61 | 321 | 11.284 | 0.1755 | Yes | ||
| 7 | AXUD1 | 369 | 10.727 | 0.1921 | Yes | ||
| 8 | TCEAL1 | 411 | 10.196 | 0.2081 | Yes | ||
| 9 | RHOB | 456 | 9.755 | 0.2231 | Yes | ||
| 10 | TSGA14 | 457 | 9.754 | 0.2405 | Yes | ||
| 11 | DUSP6 | 481 | 9.497 | 0.2563 | Yes | ||
| 12 | SLC25A12 | 497 | 9.354 | 0.2721 | Yes | ||
| 13 | PDE4D | 515 | 9.182 | 0.2876 | Yes | ||
| 14 | SCML4 | 573 | 8.556 | 0.2998 | Yes | ||
| 15 | DLX1 | 603 | 8.339 | 0.3131 | Yes | ||
| 16 | HOXB4 | 664 | 7.921 | 0.3240 | Yes | ||
| 17 | PCYT1B | 710 | 7.648 | 0.3352 | Yes | ||
| 18 | MSI2 | 786 | 7.244 | 0.3441 | Yes | ||
| 19 | THRA | 838 | 6.988 | 0.3538 | Yes | ||
| 20 | SH3BGRL | 872 | 6.868 | 0.3643 | Yes | ||
| 21 | DMD | 908 | 6.710 | 0.3743 | Yes | ||
| 22 | ATF7IP | 969 | 6.391 | 0.3825 | Yes | ||
| 23 | HOXA5 | 1022 | 6.162 | 0.3907 | Yes | ||
| 24 | RDH11 | 1043 | 6.078 | 0.4004 | Yes | ||
| 25 | FCHSD1 | 1118 | 5.758 | 0.4067 | Yes | ||
| 26 | H3F3B | 1142 | 5.607 | 0.4155 | Yes | ||
| 27 | SH3GL3 | 1266 | 5.222 | 0.4181 | Yes | ||
| 28 | SERTAD4 | 1296 | 5.117 | 0.4257 | Yes | ||
| 29 | THRAP5 | 1306 | 5.093 | 0.4343 | Yes | ||
| 30 | RRAS | 1428 | 4.766 | 0.4362 | Yes | ||
| 31 | DLGAP4 | 1481 | 4.635 | 0.4417 | Yes | ||
| 32 | TPD52 | 1486 | 4.625 | 0.4497 | Yes | ||
| 33 | DPYSL2 | 1527 | 4.522 | 0.4556 | Yes | ||
| 34 | PTEN | 1710 | 4.050 | 0.4530 | Yes | ||
| 35 | HIST1H2BM | 1759 | 3.939 | 0.4574 | Yes | ||
| 36 | KIF13A | 1830 | 3.795 | 0.4604 | Yes | ||
| 37 | HIST1H2BC | 1902 | 3.631 | 0.4630 | Yes | ||
| 38 | SREBF2 | 1968 | 3.502 | 0.4658 | Yes | ||
| 39 | NPHP4 | 2101 | 3.266 | 0.4644 | Yes | ||
| 40 | ARHGAP4 | 2163 | 3.163 | 0.4668 | Yes | ||
| 41 | NFYB | 2280 | 2.975 | 0.4658 | Yes | ||
| 42 | ETV1 | 2339 | 2.859 | 0.4677 | Yes | ||
| 43 | EYA1 | 2380 | 2.802 | 0.4706 | Yes | ||
| 44 | LPL | 2393 | 2.780 | 0.4749 | Yes | ||
| 45 | NFIA | 2432 | 2.728 | 0.4777 | Yes | ||
| 46 | GRIA3 | 2509 | 2.650 | 0.4783 | Yes | ||
| 47 | HOXB6 | 2638 | 2.506 | 0.4758 | Yes | ||
| 48 | HIST1H2BK | 2684 | 2.447 | 0.4778 | Yes | ||
| 49 | HIST1H2BB | 2693 | 2.439 | 0.4817 | Yes | ||
| 50 | HOXC5 | 2779 | 2.353 | 0.4813 | Yes | ||
| 51 | HIST1H3B | 2912 | 2.212 | 0.4781 | Yes | ||
| 52 | TBXAS1 | 2924 | 2.198 | 0.4814 | Yes | ||
| 53 | HIST1H2AH | 2934 | 2.192 | 0.4848 | Yes | ||
| 54 | RAB26 | 3096 | 2.058 | 0.4798 | No | ||
| 55 | HIST1H2AB | 3212 | 1.962 | 0.4770 | No | ||
| 56 | ALK | 3304 | 1.893 | 0.4755 | No | ||
| 57 | MAB21L2 | 3350 | 1.851 | 0.4763 | No | ||
| 58 | PIGT | 3381 | 1.827 | 0.4780 | No | ||
| 59 | TCF12 | 3555 | 1.709 | 0.4716 | No | ||
| 60 | DPYSL3 | 3678 | 1.635 | 0.4679 | No | ||
| 61 | UPK3A | 3689 | 1.628 | 0.4703 | No | ||
| 62 | BTK | 3816 | 1.546 | 0.4662 | No | ||
| 63 | HIST2H2AC | 3835 | 1.536 | 0.4680 | No | ||
| 64 | GNA14 | 3861 | 1.519 | 0.4693 | No | ||
| 65 | GAB2 | 3908 | 1.495 | 0.4695 | No | ||
| 66 | CRISP1 | 4180 | 1.343 | 0.4572 | No | ||
| 67 | HOXD8 | 4268 | 1.298 | 0.4548 | No | ||
| 68 | ARMET | 4290 | 1.282 | 0.4560 | No | ||
| 69 | TLL2 | 4291 | 1.282 | 0.4582 | No | ||
| 70 | SILV | 4451 | 1.209 | 0.4518 | No | ||
| 71 | SLC6A15 | 4535 | 1.167 | 0.4494 | No | ||
| 72 | CRYZL1 | 4666 | 1.114 | 0.4443 | No | ||
| 73 | SPRR2B | 4752 | 1.084 | 0.4416 | No | ||
| 74 | ADORA2A | 4811 | 1.057 | 0.4404 | No | ||
| 75 | NXPH1 | 4869 | 1.034 | 0.4391 | No | ||
| 76 | PLA1A | 5022 | 0.972 | 0.4326 | No | ||
| 77 | ANXA2 | 5130 | 0.934 | 0.4285 | No | ||
| 78 | BAT2 | 5144 | 0.928 | 0.4294 | No | ||
| 79 | ADORA1 | 5387 | 0.834 | 0.4178 | No | ||
| 80 | HIST1H2AC | 5485 | 0.799 | 0.4139 | No | ||
| 81 | NRL | 5514 | 0.786 | 0.4138 | No | ||
| 82 | FGF12 | 5820 | 0.676 | 0.3985 | No | ||
| 83 | TCF2 | 6021 | 0.614 | 0.3887 | No | ||
| 84 | LRRN1 | 6077 | 0.600 | 0.3868 | No | ||
| 85 | HOXB8 | 6805 | 0.381 | 0.3481 | No | ||
| 86 | ASCL3 | 6991 | 0.326 | 0.3386 | No | ||
| 87 | LYN | 7205 | 0.268 | 0.3275 | No | ||
| 88 | NEUROG1 | 7251 | 0.253 | 0.3255 | No | ||
| 89 | STX12 | 7415 | 0.204 | 0.3170 | No | ||
| 90 | POU2F3 | 7537 | 0.165 | 0.3108 | No | ||
| 91 | HPCAL4 | 7588 | 0.148 | 0.3083 | No | ||
| 92 | TMOD2 | 7826 | 0.082 | 0.2956 | No | ||
| 93 | PCDH11X | 7915 | 0.057 | 0.2909 | No | ||
| 94 | HIST1H3C | 7999 | 0.031 | 0.2865 | No | ||
| 95 | VSNL1 | 8042 | 0.018 | 0.2843 | No | ||
| 96 | CNNM4 | 8072 | 0.011 | 0.2827 | No | ||
| 97 | PFKFB1 | 8179 | -0.019 | 0.2770 | No | ||
| 98 | ARF6 | 8478 | -0.100 | 0.2610 | No | ||
| 99 | CLCA3 | 8908 | -0.207 | 0.2381 | No | ||
| 100 | SFRS14 | 9136 | -0.260 | 0.2262 | No | ||
| 101 | PRRX1 | 9347 | -0.312 | 0.2154 | No | ||
| 102 | PAX6 | 9397 | -0.326 | 0.2133 | No | ||
| 103 | HOXA10 | 9464 | -0.340 | 0.2104 | No | ||
| 104 | NOS1 | 9465 | -0.340 | 0.2110 | No | ||
| 105 | LIPG | 9472 | -0.342 | 0.2112 | No | ||
| 106 | SFRP1 | 9743 | -0.408 | 0.1973 | No | ||
| 107 | SMPX | 9926 | -0.459 | 0.1883 | No | ||
| 108 | IRX4 | 10079 | -0.499 | 0.1809 | No | ||
| 109 | ALDH1A1 | 10135 | -0.511 | 0.1788 | No | ||
| 110 | FZD4 | 10175 | -0.521 | 0.1777 | No | ||
| 111 | PHOX2B | 10261 | -0.547 | 0.1740 | No | ||
| 112 | HIST1H2AA | 10276 | -0.551 | 0.1742 | No | ||
| 113 | WNT6 | 10421 | -0.586 | 0.1675 | No | ||
| 114 | OTX2 | 10429 | -0.588 | 0.1681 | No | ||
| 115 | UBE2S | 10534 | -0.611 | 0.1636 | No | ||
| 116 | FOXP1 | 10892 | -0.694 | 0.1455 | No | ||
| 117 | SRF | 10949 | -0.705 | 0.1437 | No | ||
| 118 | RPP21 | 11030 | -0.723 | 0.1406 | No | ||
| 119 | PPP2R5C | 11124 | -0.744 | 0.1369 | No | ||
| 120 | EGR2 | 11225 | -0.769 | 0.1329 | No | ||
| 121 | SLITRK6 | 11372 | -0.806 | 0.1264 | No | ||
| 122 | HOXD11 | 11519 | -0.847 | 0.1200 | No | ||
| 123 | MGAT5B | 11781 | -0.911 | 0.1074 | No | ||
| 124 | NDNL2 | 11883 | -0.938 | 0.1036 | No | ||
| 125 | GPR85 | 11894 | -0.942 | 0.1048 | No | ||
| 126 | FSTL5 | 12269 | -1.043 | 0.0863 | No | ||
| 127 | NRXN3 | 12368 | -1.068 | 0.0829 | No | ||
| 128 | GPR4 | 12497 | -1.108 | 0.0780 | No | ||
| 129 | MRAS | 12558 | -1.126 | 0.0767 | No | ||
| 130 | SYNPR | 12651 | -1.156 | 0.0738 | No | ||
| 131 | REL | 12776 | -1.191 | 0.0692 | No | ||
| 132 | SCOC | 12807 | -1.200 | 0.0697 | No | ||
| 133 | BDNF | 12829 | -1.208 | 0.0707 | No | ||
| 134 | TCERG1L | 12976 | -1.255 | 0.0651 | No | ||
| 135 | GPM6A | 12986 | -1.258 | 0.0668 | No | ||
| 136 | ZHX2 | 13005 | -1.264 | 0.0681 | No | ||
| 137 | MTUS1 | 13089 | -1.287 | 0.0659 | No | ||
| 138 | RARB | 13124 | -1.297 | 0.0664 | No | ||
| 139 | PTHLH | 13276 | -1.347 | 0.0606 | No | ||
| 140 | ITSN1 | 13278 | -1.348 | 0.0629 | No | ||
| 141 | CDX4 | 13305 | -1.356 | 0.0639 | No | ||
| 142 | CDX2 | 13481 | -1.416 | 0.0570 | No | ||
| 143 | ACCN1 | 13522 | -1.431 | 0.0574 | No | ||
| 144 | FOXP2 | 13796 | -1.525 | 0.0453 | No | ||
| 145 | HIST1H2BA | 13834 | -1.541 | 0.0460 | No | ||
| 146 | SFRP2 | 13861 | -1.556 | 0.0474 | No | ||
| 147 | CSF3 | 13921 | -1.574 | 0.0470 | No | ||
| 148 | EHF | 14062 | -1.624 | 0.0423 | No | ||
| 149 | EAF2 | 14105 | -1.644 | 0.0430 | No | ||
| 150 | FBXO24 | 14147 | -1.658 | 0.0437 | No | ||
| 151 | FGF20 | 14237 | -1.693 | 0.0419 | No | ||
| 152 | POU3F4 | 14260 | -1.702 | 0.0437 | No | ||
| 153 | MAP2K6 | 14286 | -1.709 | 0.0454 | No | ||
| 154 | LHX6 | 14416 | -1.771 | 0.0416 | No | ||
| 155 | PRICKLE2 | 14657 | -1.882 | 0.0319 | No | ||
| 156 | FGF14 | 14660 | -1.883 | 0.0352 | No | ||
| 157 | PRDM1 | 14786 | -1.938 | 0.0319 | No | ||
| 158 | NRXN1 | 14903 | -2.001 | 0.0291 | No | ||
| 159 | MID1 | 14907 | -2.003 | 0.0325 | No | ||
| 160 | TRERF1 | 14999 | -2.046 | 0.0313 | No | ||
| 161 | SP6 | 15189 | -2.150 | 0.0248 | No | ||
| 162 | COL25A1 | 15294 | -2.213 | 0.0232 | No | ||
| 163 | HOXB3 | 15359 | -2.260 | 0.0237 | No | ||
| 164 | LRCH4 | 15409 | -2.286 | 0.0251 | No | ||
| 165 | SEMA6C | 15444 | -2.317 | 0.0274 | No | ||
| 166 | PHF6 | 15599 | -2.417 | 0.0234 | No | ||
| 167 | DAPK3 | 15631 | -2.441 | 0.0261 | No | ||
| 168 | PCDH8 | 15668 | -2.466 | 0.0285 | No | ||
| 169 | GALNT1 | 15802 | -2.594 | 0.0259 | No | ||
| 170 | ATP2A2 | 16007 | -2.790 | 0.0198 | No | ||
| 171 | NIN | 16014 | -2.795 | 0.0245 | No | ||
| 172 | ISL1 | 16376 | -3.139 | 0.0105 | No | ||
| 173 | E2F3 | 16497 | -3.306 | 0.0099 | No | ||
| 174 | RTTN | 16715 | -3.619 | 0.0046 | No | ||
| 175 | TRAF3 | 16727 | -3.648 | 0.0105 | No | ||
| 176 | BCL2 | 16768 | -3.720 | 0.0150 | No | ||
| 177 | LDB2 | 16861 | -3.859 | 0.0169 | No | ||
| 178 | DSCR1 | 16867 | -3.870 | 0.0235 | No | ||
| 179 | CDK2 | 16879 | -3.891 | 0.0299 | No | ||
| 180 | NRAS | 17023 | -4.143 | 0.0295 | No | ||
| 181 | KCNN3 | 17310 | -4.792 | 0.0226 | No | ||
| 182 | CD86 | 17499 | -5.321 | 0.0219 | No | ||
| 183 | SFRS2 | 17611 | -5.660 | 0.0260 | No | ||
| 184 | PIM2 | 17951 | -7.139 | 0.0203 | No | ||
| 185 | TSNAX | 18223 | -8.802 | 0.0213 | No |