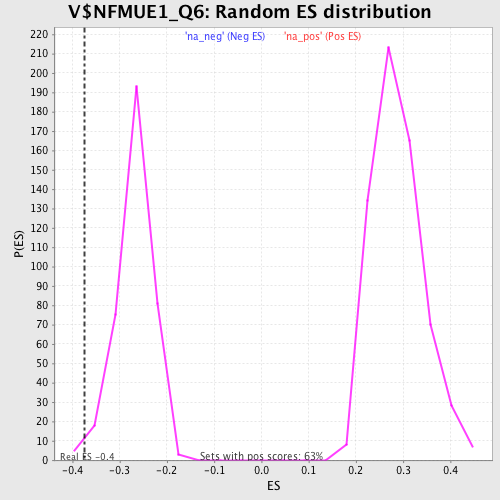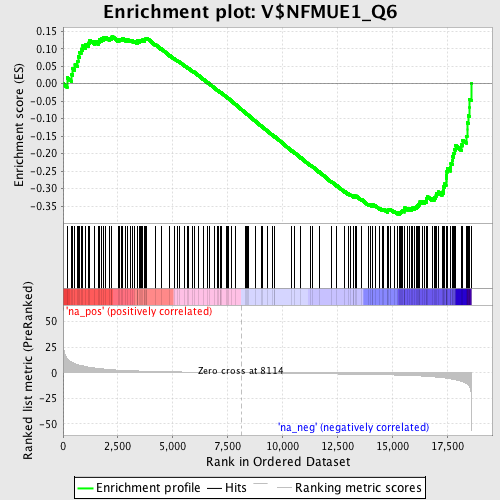
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$NFMUE1_Q6 |
| Enrichment Score (ES) | -0.37409693 |
| Normalized Enrichment Score (NES) | -1.399009 |
| Nominal p-value | 0.013333334 |
| FDR q-value | 0.18557799 |
| FWER p-Value | 1.0 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | APBB1 | 182 | 13.954 | 0.0173 | No | ||
| 2 | MAML1 | 394 | 10.451 | 0.0262 | No | ||
| 3 | PIAS3 | 433 | 9.998 | 0.0437 | No | ||
| 4 | H1F0 | 540 | 8.900 | 0.0553 | No | ||
| 5 | EPC1 | 657 | 7.949 | 0.0644 | No | ||
| 6 | RIOK3 | 685 | 7.811 | 0.0782 | No | ||
| 7 | TCF4 | 752 | 7.435 | 0.0891 | No | ||
| 8 | ASB2 | 820 | 7.056 | 0.0992 | No | ||
| 9 | DGCR2 | 899 | 6.743 | 0.1081 | No | ||
| 10 | DDX6 | 1032 | 6.123 | 0.1129 | No | ||
| 11 | RAB1A | 1167 | 5.520 | 0.1164 | No | ||
| 12 | RAB22A | 1214 | 5.380 | 0.1244 | No | ||
| 13 | NDST2 | 1430 | 4.751 | 0.1220 | No | ||
| 14 | UTX | 1610 | 4.282 | 0.1206 | No | ||
| 15 | MYNN | 1650 | 4.190 | 0.1267 | No | ||
| 16 | RALA | 1758 | 3.940 | 0.1286 | No | ||
| 17 | CHD2 | 1845 | 3.767 | 0.1312 | No | ||
| 18 | XPR1 | 1941 | 3.557 | 0.1330 | No | ||
| 19 | WASL | 2116 | 3.241 | 0.1299 | No | ||
| 20 | ZADH2 | 2195 | 3.119 | 0.1317 | No | ||
| 21 | ZFX | 2227 | 3.071 | 0.1361 | No | ||
| 22 | EXT1 | 2517 | 2.642 | 0.1255 | No | ||
| 23 | POLK | 2584 | 2.567 | 0.1270 | No | ||
| 24 | UQCRH | 2674 | 2.461 | 0.1269 | No | ||
| 25 | STAG1 | 2703 | 2.430 | 0.1301 | No | ||
| 26 | POU2F1 | 2859 | 2.270 | 0.1262 | No | ||
| 27 | BMI1 | 2943 | 2.184 | 0.1259 | No | ||
| 28 | ZIC3 | 3056 | 2.089 | 0.1239 | No | ||
| 29 | RBM4 | 3145 | 2.017 | 0.1231 | No | ||
| 30 | RNF26 | 3260 | 1.931 | 0.1207 | No | ||
| 31 | EEF1A1 | 3386 | 1.825 | 0.1175 | No | ||
| 32 | RBM5 | 3387 | 1.824 | 0.1210 | No | ||
| 33 | ATP6V0C | 3393 | 1.821 | 0.1243 | No | ||
| 34 | ZFP37 | 3497 | 1.745 | 0.1221 | No | ||
| 35 | SUV39H2 | 3519 | 1.730 | 0.1243 | No | ||
| 36 | RFX3 | 3576 | 1.700 | 0.1246 | No | ||
| 37 | PTBP2 | 3624 | 1.670 | 0.1253 | No | ||
| 38 | VAMP2 | 3727 | 1.606 | 0.1229 | No | ||
| 39 | CBX5 | 3730 | 1.605 | 0.1259 | No | ||
| 40 | RAC1 | 3748 | 1.593 | 0.1281 | No | ||
| 41 | PHF15 | 3782 | 1.567 | 0.1294 | No | ||
| 42 | ARF1 | 3817 | 1.546 | 0.1306 | No | ||
| 43 | ATF1 | 4203 | 1.334 | 0.1123 | No | ||
| 44 | PICALM | 4468 | 1.201 | 0.1003 | No | ||
| 45 | AP3D1 | 4866 | 1.035 | 0.0808 | No | ||
| 46 | SLC39A9 | 5069 | 0.955 | 0.0717 | No | ||
| 47 | UBE2D3 | 5234 | 0.895 | 0.0646 | No | ||
| 48 | NSD1 | 5307 | 0.864 | 0.0624 | No | ||
| 49 | BRD3 | 5533 | 0.777 | 0.0517 | No | ||
| 50 | OLFML3 | 5689 | 0.716 | 0.0447 | No | ||
| 51 | STRN4 | 5728 | 0.705 | 0.0440 | No | ||
| 52 | CSAD | 5897 | 0.649 | 0.0362 | No | ||
| 53 | ASH1L | 5969 | 0.632 | 0.0336 | No | ||
| 54 | ATP5H | 6163 | 0.573 | 0.0242 | No | ||
| 55 | HNRPD | 6399 | 0.500 | 0.0125 | No | ||
| 56 | JARID1C | 6595 | 0.443 | 0.0028 | No | ||
| 57 | USP48 | 6675 | 0.422 | -0.0007 | No | ||
| 58 | CPSF2 | 6887 | 0.358 | -0.0114 | No | ||
| 59 | CPNE1 | 7032 | 0.314 | -0.0186 | No | ||
| 60 | PHF8 | 7088 | 0.298 | -0.0210 | No | ||
| 61 | TSC1 | 7178 | 0.276 | -0.0253 | No | ||
| 62 | TIA1 | 7194 | 0.270 | -0.0256 | No | ||
| 63 | FKRP | 7210 | 0.267 | -0.0259 | No | ||
| 64 | HOXA2 | 7432 | 0.198 | -0.0375 | No | ||
| 65 | NCOR1 | 7474 | 0.186 | -0.0393 | No | ||
| 66 | CD3E | 7530 | 0.167 | -0.0420 | No | ||
| 67 | PCF11 | 7692 | 0.121 | -0.0505 | No | ||
| 68 | RPS27 | 7864 | 0.070 | -0.0596 | No | ||
| 69 | RPL35A | 8307 | -0.058 | -0.0834 | No | ||
| 70 | PCBP4 | 8351 | -0.070 | -0.0856 | No | ||
| 71 | NFYA | 8419 | -0.085 | -0.0891 | No | ||
| 72 | HMGB3 | 8445 | -0.091 | -0.0903 | No | ||
| 73 | NEUROD2 | 8773 | -0.174 | -0.1077 | No | ||
| 74 | REST | 9026 | -0.236 | -0.1209 | No | ||
| 75 | PABPN1 | 9064 | -0.243 | -0.1224 | No | ||
| 76 | CD4 | 9308 | -0.306 | -0.1350 | No | ||
| 77 | DEAF1 | 9525 | -0.353 | -0.1460 | No | ||
| 78 | CCAR1 | 9652 | -0.383 | -0.1521 | No | ||
| 79 | PIGN | 9653 | -0.383 | -0.1513 | No | ||
| 80 | OTX2 | 10429 | -0.588 | -0.1922 | No | ||
| 81 | COL4A3BP | 10561 | -0.617 | -0.1980 | No | ||
| 82 | SFPQ | 10840 | -0.683 | -0.2118 | No | ||
| 83 | CSTF2T | 11260 | -0.777 | -0.2330 | No | ||
| 84 | SRRM1 | 11348 | -0.799 | -0.2361 | No | ||
| 85 | ATP5G2 | 11669 | -0.882 | -0.2517 | No | ||
| 86 | NFYC | 12241 | -1.035 | -0.2807 | No | ||
| 87 | PAX2 | 12472 | -1.102 | -0.2910 | No | ||
| 88 | PHC3 | 12839 | -1.211 | -0.3085 | No | ||
| 89 | CLSTN1 | 12984 | -1.258 | -0.3138 | No | ||
| 90 | HNRPA1 | 13113 | -1.295 | -0.3182 | No | ||
| 91 | CDH23 | 13241 | -1.336 | -0.3225 | No | ||
| 92 | BCL11B | 13246 | -1.337 | -0.3201 | No | ||
| 93 | CNOT3 | 13346 | -1.372 | -0.3228 | No | ||
| 94 | APBB3 | 13366 | -1.378 | -0.3212 | No | ||
| 95 | RBM12 | 13612 | -1.464 | -0.3316 | No | ||
| 96 | SNAP25 | 13920 | -1.574 | -0.3451 | No | ||
| 97 | NKIRAS2 | 14002 | -1.605 | -0.3464 | No | ||
| 98 | MARK3 | 14089 | -1.637 | -0.3479 | No | ||
| 99 | ACTR8 | 14096 | -1.641 | -0.3450 | No | ||
| 100 | E4F1 | 14219 | -1.685 | -0.3483 | No | ||
| 101 | PSMD8 | 14434 | -1.779 | -0.3565 | No | ||
| 102 | PIGL | 14539 | -1.829 | -0.3585 | No | ||
| 103 | PRKCSH | 14615 | -1.861 | -0.3590 | No | ||
| 104 | DHX15 | 14785 | -1.937 | -0.3644 | No | ||
| 105 | RPL30 | 14798 | -1.947 | -0.3612 | No | ||
| 106 | PRPS1 | 14849 | -1.970 | -0.3601 | No | ||
| 107 | MID1 | 14907 | -2.003 | -0.3593 | No | ||
| 108 | RBM3 | 15094 | -2.095 | -0.3653 | No | ||
| 109 | ARNT | 15236 | -2.177 | -0.3687 | No | ||
| 110 | PSMB5 | 15337 | -2.243 | -0.3697 | Yes | ||
| 111 | ATP5F1 | 15369 | -2.265 | -0.3670 | Yes | ||
| 112 | TNRC15 | 15421 | -2.295 | -0.3653 | Yes | ||
| 113 | SLC35A4 | 15453 | -2.320 | -0.3624 | Yes | ||
| 114 | PRPSAP1 | 15539 | -2.379 | -0.3624 | Yes | ||
| 115 | CLK1 | 15562 | -2.391 | -0.3590 | Yes | ||
| 116 | ARCN1 | 15576 | -2.399 | -0.3550 | Yes | ||
| 117 | USF1 | 15698 | -2.488 | -0.3567 | Yes | ||
| 118 | SMCR8 | 15805 | -2.595 | -0.3574 | Yes | ||
| 119 | ATF4 | 15892 | -2.676 | -0.3568 | Yes | ||
| 120 | YY1 | 15942 | -2.726 | -0.3542 | Yes | ||
| 121 | OSR1 | 16036 | -2.815 | -0.3537 | Yes | ||
| 122 | FASTK | 16114 | -2.881 | -0.3523 | Yes | ||
| 123 | TOP3A | 16142 | -2.915 | -0.3481 | Yes | ||
| 124 | PPP1R15B | 16200 | -2.969 | -0.3454 | Yes | ||
| 125 | ODF2 | 16236 | -3.003 | -0.3414 | Yes | ||
| 126 | UBE4B | 16264 | -3.035 | -0.3370 | Yes | ||
| 127 | MAEA | 16388 | -3.151 | -0.3375 | Yes | ||
| 128 | RAD23A | 16483 | -3.284 | -0.3362 | Yes | ||
| 129 | PBX3 | 16555 | -3.374 | -0.3335 | Yes | ||
| 130 | RPAP1 | 16573 | -3.402 | -0.3278 | Yes | ||
| 131 | BCLAF1 | 16609 | -3.463 | -0.3229 | Yes | ||
| 132 | SFRS10 | 16834 | -3.818 | -0.3276 | Yes | ||
| 133 | THAP1 | 16932 | -3.974 | -0.3251 | Yes | ||
| 134 | PPP1CC | 16994 | -4.092 | -0.3205 | Yes | ||
| 135 | NCDN | 17016 | -4.130 | -0.3136 | Yes | ||
| 136 | ENSA | 17094 | -4.295 | -0.3094 | Yes | ||
| 137 | EIF4G2 | 17271 | -4.699 | -0.3098 | Yes | ||
| 138 | IRAK1 | 17337 | -4.869 | -0.3038 | Yes | ||
| 139 | E2F6 | 17351 | -4.899 | -0.2950 | Yes | ||
| 140 | GNPAT | 17375 | -4.977 | -0.2865 | Yes | ||
| 141 | PPARBP | 17456 | -5.199 | -0.2807 | Yes | ||
| 142 | SNRPF | 17457 | -5.204 | -0.2706 | Yes | ||
| 143 | EIF5 | 17471 | -5.251 | -0.2611 | Yes | ||
| 144 | DNAJC7 | 17488 | -5.299 | -0.2516 | Yes | ||
| 145 | RPA2 | 17511 | -5.355 | -0.2424 | Yes | ||
| 146 | NASP | 17651 | -5.801 | -0.2386 | Yes | ||
| 147 | EIF3S10 | 17674 | -5.899 | -0.2283 | Yes | ||
| 148 | CUGBP1 | 17743 | -6.188 | -0.2199 | Yes | ||
| 149 | VDAC2 | 17759 | -6.247 | -0.2086 | Yes | ||
| 150 | SMYD5 | 17806 | -6.484 | -0.1984 | Yes | ||
| 151 | ERH | 17828 | -6.552 | -0.1868 | Yes | ||
| 152 | UBE2J2 | 17888 | -6.823 | -0.1767 | Yes | ||
| 153 | NACA | 18147 | -8.210 | -0.1747 | Yes | ||
| 154 | WBSCR16 | 18213 | -8.645 | -0.1614 | Yes | ||
| 155 | PPRC1 | 18372 | -10.163 | -0.1502 | Yes | ||
| 156 | DAP3 | 18412 | -10.667 | -0.1315 | Yes | ||
| 157 | ADK | 18413 | -10.674 | -0.1107 | Yes | ||
| 158 | CCNE1 | 18465 | -11.544 | -0.0910 | Yes | ||
| 159 | RBM19 | 18510 | -12.702 | -0.0686 | Yes | ||
| 160 | MRPL38 | 18515 | -12.810 | -0.0439 | Yes | ||
| 161 | CCND1 | 18607 | -25.315 | 0.0005 | Yes |