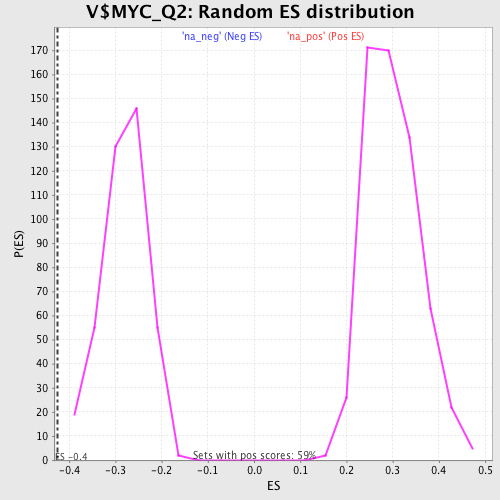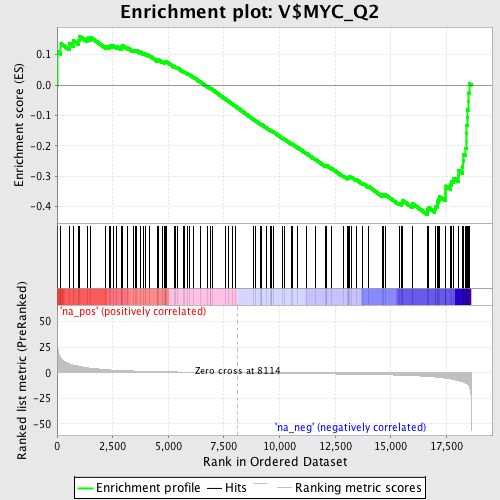
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$MYC_Q2 |
| Enrichment Score (ES) | -0.426159 |
| Normalized Enrichment Score (NES) | -1.5170351 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.10019235 |
| FWER p-Value | 0.932 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | FXYD6 | 3 | 46.001 | 0.1111 | No | ||
| 2 | BCL9L | 173 | 14.167 | 0.1363 | No | ||
| 3 | PSEN2 | 558 | 8.737 | 0.1367 | No | ||
| 4 | KRTCAP2 | 726 | 7.549 | 0.1459 | No | ||
| 5 | ATF7IP | 969 | 6.391 | 0.1483 | No | ||
| 6 | PFKFB3 | 1011 | 6.224 | 0.1611 | No | ||
| 7 | COMMD3 | 1348 | 4.966 | 0.1550 | No | ||
| 8 | CDC14A | 1503 | 4.558 | 0.1577 | No | ||
| 9 | RAB3IL1 | 2188 | 3.132 | 0.1283 | No | ||
| 10 | USP15 | 2343 | 2.855 | 0.1269 | No | ||
| 11 | USP2 | 2397 | 2.776 | 0.1307 | No | ||
| 12 | PTPRF | 2528 | 2.629 | 0.1300 | No | ||
| 13 | RRAGC | 2686 | 2.446 | 0.1275 | No | ||
| 14 | LTBR | 2894 | 2.237 | 0.1217 | No | ||
| 15 | TMEM24 | 2897 | 2.234 | 0.1270 | No | ||
| 16 | H3F3A | 2950 | 2.175 | 0.1294 | No | ||
| 17 | ESRRA | 3153 | 2.011 | 0.1234 | No | ||
| 18 | COL2A1 | 3416 | 1.804 | 0.1136 | No | ||
| 19 | AP3M1 | 3504 | 1.739 | 0.1131 | No | ||
| 20 | NIT1 | 3565 | 1.705 | 0.1140 | No | ||
| 21 | CBX5 | 3730 | 1.605 | 0.1090 | No | ||
| 22 | ALDH3B1 | 3870 | 1.515 | 0.1051 | No | ||
| 23 | TDRD1 | 3988 | 1.444 | 0.1023 | No | ||
| 24 | CHD4 | 4159 | 1.353 | 0.0964 | No | ||
| 25 | FCHSD2 | 4493 | 1.188 | 0.0813 | No | ||
| 26 | CTBP2 | 4514 | 1.176 | 0.0830 | No | ||
| 27 | SLC6A15 | 4535 | 1.167 | 0.0848 | No | ||
| 28 | RUSC1 | 4758 | 1.081 | 0.0754 | No | ||
| 29 | APOA5 | 4820 | 1.054 | 0.0746 | No | ||
| 30 | SIRT1 | 4852 | 1.042 | 0.0755 | No | ||
| 31 | GPD1 | 4868 | 1.034 | 0.0772 | No | ||
| 32 | HMGN2 | 4895 | 1.023 | 0.0782 | No | ||
| 33 | CTSF | 5272 | 0.879 | 0.0600 | No | ||
| 34 | FOXD3 | 5329 | 0.855 | 0.0591 | No | ||
| 35 | RPL22 | 5403 | 0.829 | 0.0571 | No | ||
| 36 | LIN28 | 5677 | 0.720 | 0.0441 | No | ||
| 37 | FADS3 | 5746 | 0.699 | 0.0421 | No | ||
| 38 | NKX2-3 | 5870 | 0.658 | 0.0371 | No | ||
| 39 | ILK | 5946 | 0.637 | 0.0346 | No | ||
| 40 | LRP8 | 6151 | 0.578 | 0.0249 | No | ||
| 41 | SLC1A7 | 6422 | 0.494 | 0.0115 | No | ||
| 42 | TRIM46 | 6766 | 0.392 | -0.0060 | No | ||
| 43 | ALX4 | 6878 | 0.361 | -0.0112 | No | ||
| 44 | PITX3 | 6907 | 0.354 | -0.0118 | No | ||
| 45 | HPCA | 6990 | 0.326 | -0.0155 | No | ||
| 46 | HPCAL4 | 7588 | 0.148 | -0.0474 | No | ||
| 47 | LHX9 | 7703 | 0.116 | -0.0533 | No | ||
| 48 | CHRM1 | 7872 | 0.069 | -0.0622 | No | ||
| 49 | B4GALT2 | 8021 | 0.026 | -0.0701 | No | ||
| 50 | OPRD1 | 8835 | -0.188 | -0.1136 | No | ||
| 51 | IPO13 | 8929 | -0.212 | -0.1181 | No | ||
| 52 | HOXC13 | 9157 | -0.267 | -0.1298 | No | ||
| 53 | WEE1 | 9178 | -0.271 | -0.1302 | No | ||
| 54 | TGFB2 | 9206 | -0.277 | -0.1310 | No | ||
| 55 | PAX6 | 9397 | -0.326 | -0.1405 | No | ||
| 56 | SC5DL | 9600 | -0.372 | -0.1505 | No | ||
| 57 | TAGLN2 | 9642 | -0.381 | -0.1518 | No | ||
| 58 | NRIP3 | 9650 | -0.382 | -0.1512 | No | ||
| 59 | PLA2G4A | 9733 | -0.406 | -0.1547 | No | ||
| 60 | POGK | 10138 | -0.512 | -0.1753 | No | ||
| 61 | NR0B2 | 10212 | -0.531 | -0.1779 | No | ||
| 62 | STMN1 | 10556 | -0.615 | -0.1950 | No | ||
| 63 | UBXD3 | 10566 | -0.619 | -0.1940 | No | ||
| 64 | HNRPF | 10822 | -0.678 | -0.2061 | No | ||
| 65 | CNNM1 | 11197 | -0.764 | -0.2245 | No | ||
| 66 | CGN | 11600 | -0.868 | -0.2441 | No | ||
| 67 | B3GALT2 | 12064 | -0.987 | -0.2668 | No | ||
| 68 | TAF6L | 12096 | -0.997 | -0.2660 | No | ||
| 69 | SLC16A1 | 12126 | -1.006 | -0.2652 | No | ||
| 70 | AGMAT | 12352 | -1.064 | -0.2747 | No | ||
| 71 | RTN4RL2 | 12867 | -1.221 | -0.2996 | No | ||
| 72 | FOSL1 | 13050 | -1.278 | -0.3063 | No | ||
| 73 | SCYL1 | 13091 | -1.288 | -0.3054 | No | ||
| 74 | HNRPA1 | 13113 | -1.295 | -0.3034 | No | ||
| 75 | LMX1A | 13122 | -1.297 | -0.3007 | No | ||
| 76 | HOXC12 | 13220 | -1.328 | -0.3027 | No | ||
| 77 | TESK2 | 13441 | -1.402 | -0.3112 | No | ||
| 78 | RFX5 | 13740 | -1.502 | -0.3237 | No | ||
| 79 | FXYD2 | 13981 | -1.598 | -0.3328 | No | ||
| 80 | ATP6V0B | 14612 | -1.861 | -0.3623 | No | ||
| 81 | SYT6 | 14656 | -1.881 | -0.3601 | No | ||
| 82 | RORC | 14765 | -1.929 | -0.3613 | No | ||
| 83 | ATP5F1 | 15369 | -2.265 | -0.3884 | No | ||
| 84 | TIAL1 | 15496 | -2.348 | -0.3895 | No | ||
| 85 | IPO7 | 15529 | -2.373 | -0.3855 | No | ||
| 86 | POU3F1 | 15543 | -2.381 | -0.3805 | No | ||
| 87 | FABP3 | 15971 | -2.751 | -0.3969 | No | ||
| 88 | CENTB5 | 15977 | -2.758 | -0.3905 | No | ||
| 89 | IL15RA | 16638 | -3.510 | -0.4177 | Yes | ||
| 90 | B3GALT6 | 16654 | -3.530 | -0.4099 | Yes | ||
| 91 | BHLHB3 | 16712 | -3.613 | -0.4043 | Yes | ||
| 92 | CFL1 | 16985 | -4.061 | -0.4092 | Yes | ||
| 93 | NRAS | 17023 | -4.143 | -0.4011 | Yes | ||
| 94 | PFDN2 | 17082 | -4.269 | -0.3939 | Yes | ||
| 95 | TFB2M | 17101 | -4.311 | -0.3845 | Yes | ||
| 96 | ZZZ3 | 17143 | -4.402 | -0.3760 | Yes | ||
| 97 | RHEBL1 | 17192 | -4.526 | -0.3677 | Yes | ||
| 98 | ARL3 | 17448 | -5.159 | -0.3690 | Yes | ||
| 99 | GTF2H1 | 17462 | -5.217 | -0.3571 | Yes | ||
| 100 | EIF4B | 17470 | -5.251 | -0.3448 | Yes | ||
| 101 | AMPD2 | 17478 | -5.275 | -0.3324 | Yes | ||
| 102 | EIF3S10 | 17674 | -5.899 | -0.3286 | Yes | ||
| 103 | IVNS1ABP | 17729 | -6.121 | -0.3167 | Yes | ||
| 104 | PA2G4 | 17835 | -6.589 | -0.3065 | Yes | ||
| 105 | PABPC4 | 18028 | -7.556 | -0.2986 | Yes | ||
| 106 | COPS7A | 18050 | -7.721 | -0.2810 | Yes | ||
| 107 | SFXN2 | 18227 | -8.818 | -0.2692 | Yes | ||
| 108 | TIMM10 | 18244 | -8.958 | -0.2484 | Yes | ||
| 109 | SHMT2 | 18277 | -9.181 | -0.2279 | Yes | ||
| 110 | PPRC1 | 18372 | -10.163 | -0.2084 | Yes | ||
| 111 | AK2 | 18405 | -10.572 | -0.1846 | Yes | ||
| 112 | LDHA | 18407 | -10.608 | -0.1590 | Yes | ||
| 113 | ADK | 18413 | -10.674 | -0.1334 | Yes | ||
| 114 | CSDA | 18435 | -10.982 | -0.1080 | Yes | ||
| 115 | NUDC | 18442 | -11.093 | -0.0815 | Yes | ||
| 116 | ATAD3A | 18475 | -11.799 | -0.0546 | Yes | ||
| 117 | EBNA1BP2 | 18478 | -11.855 | -0.0261 | Yes | ||
| 118 | FKBP11 | 18539 | -13.834 | 0.0042 | Yes |