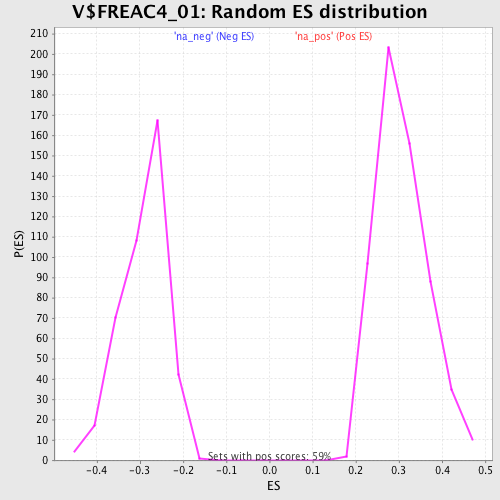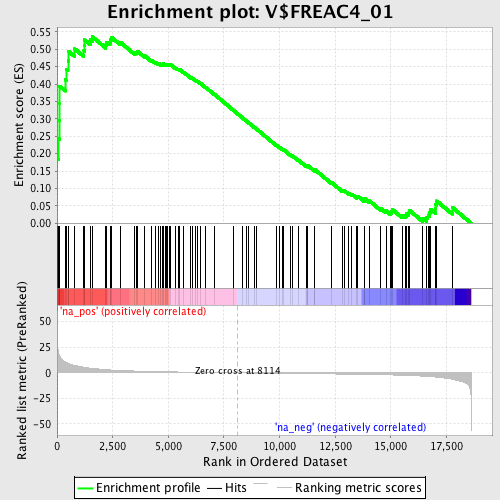
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FREAC4_01 |
| Enrichment Score (ES) | 0.5362675 |
| Normalized Enrichment Score (NES) | 1.7507871 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.045958225 |
| FWER p-Value | 0.257 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 60.375 | 0.1860 | Yes | ||
| 2 | TCF7L2 | 59 | 19.757 | 0.2438 | Yes | ||
| 3 | CDKN1A | 100 | 17.229 | 0.2947 | Yes | ||
| 4 | LEF1 | 114 | 16.418 | 0.3446 | Yes | ||
| 5 | GFRA1 | 120 | 16.047 | 0.3938 | Yes | ||
| 6 | BTBD3 | 378 | 10.592 | 0.4125 | Yes | ||
| 7 | ERG | 405 | 10.291 | 0.4428 | Yes | ||
| 8 | MBNL2 | 490 | 9.423 | 0.4673 | Yes | ||
| 9 | ETHE1 | 528 | 9.010 | 0.4931 | Yes | ||
| 10 | DUSP1 | 797 | 7.185 | 0.5008 | Yes | ||
| 11 | CTCF | 1205 | 5.422 | 0.4955 | Yes | ||
| 12 | IGF1 | 1226 | 5.346 | 0.5109 | Yes | ||
| 13 | PLS3 | 1233 | 5.322 | 0.5270 | Yes | ||
| 14 | TLE3 | 1498 | 4.578 | 0.5268 | Yes | ||
| 15 | KBTBD2 | 1575 | 4.401 | 0.5363 | Yes | ||
| 16 | ZADH2 | 2195 | 3.119 | 0.5125 | No | ||
| 17 | ZFX | 2227 | 3.071 | 0.5203 | No | ||
| 18 | EYA1 | 2380 | 2.802 | 0.5207 | No | ||
| 19 | CAMK1D | 2410 | 2.758 | 0.5276 | No | ||
| 20 | PELI2 | 2448 | 2.708 | 0.5340 | No | ||
| 21 | CALD1 | 2832 | 2.299 | 0.5204 | No | ||
| 22 | UBE2H | 3495 | 1.747 | 0.4900 | No | ||
| 23 | SLC24A2 | 3568 | 1.702 | 0.4914 | No | ||
| 24 | TCF1 | 3628 | 1.668 | 0.4933 | No | ||
| 25 | FOXB1 | 3943 | 1.470 | 0.4809 | No | ||
| 26 | PITX2 | 4241 | 1.312 | 0.4689 | No | ||
| 27 | OTX1 | 4443 | 1.214 | 0.4618 | No | ||
| 28 | HOXA11 | 4567 | 1.154 | 0.4587 | No | ||
| 29 | PUM2 | 4657 | 1.119 | 0.4574 | No | ||
| 30 | ULK1 | 4747 | 1.086 | 0.4559 | No | ||
| 31 | SYT10 | 4760 | 1.080 | 0.4586 | No | ||
| 32 | OTOP3 | 4861 | 1.039 | 0.4564 | No | ||
| 33 | ROR1 | 4908 | 1.019 | 0.4570 | No | ||
| 34 | VIT | 4980 | 0.986 | 0.4562 | No | ||
| 35 | CITED2 | 5036 | 0.967 | 0.4562 | No | ||
| 36 | HOXC6 | 5103 | 0.941 | 0.4556 | No | ||
| 37 | FOXD3 | 5329 | 0.855 | 0.4461 | No | ||
| 38 | PER2 | 5436 | 0.818 | 0.4429 | No | ||
| 39 | MSC | 5509 | 0.788 | 0.4414 | No | ||
| 40 | PNRC1 | 5690 | 0.715 | 0.4339 | No | ||
| 41 | CD109 | 6006 | 0.617 | 0.4188 | No | ||
| 42 | PHEX | 6072 | 0.602 | 0.4171 | No | ||
| 43 | FGF9 | 6230 | 0.552 | 0.4104 | No | ||
| 44 | TRPS1 | 6309 | 0.529 | 0.4078 | No | ||
| 45 | LOX | 6438 | 0.489 | 0.4024 | No | ||
| 46 | GABRR1 | 6691 | 0.417 | 0.3901 | No | ||
| 47 | TLX1 | 7060 | 0.306 | 0.3711 | No | ||
| 48 | HOXD9 | 7932 | 0.052 | 0.3243 | No | ||
| 49 | ODZ1 | 8348 | -0.070 | 0.3021 | No | ||
| 50 | NR4A1 | 8495 | -0.105 | 0.2945 | No | ||
| 51 | PURA | 8623 | -0.136 | 0.2881 | No | ||
| 52 | SPRY4 | 8864 | -0.194 | 0.2757 | No | ||
| 53 | MYH2 | 8972 | -0.222 | 0.2706 | No | ||
| 54 | IBSP | 9846 | -0.436 | 0.2249 | No | ||
| 55 | OSR2 | 10004 | -0.478 | 0.2178 | No | ||
| 56 | CLDN18 | 10130 | -0.510 | 0.2127 | No | ||
| 57 | TGFB3 | 10184 | -0.524 | 0.2114 | No | ||
| 58 | COLEC12 | 10477 | -0.597 | 0.1975 | No | ||
| 59 | UBXD3 | 10566 | -0.619 | 0.1947 | No | ||
| 60 | PTHR1 | 10845 | -0.683 | 0.1818 | No | ||
| 61 | PLUNC | 11213 | -0.767 | 0.1643 | No | ||
| 62 | SLC34A3 | 11261 | -0.777 | 0.1642 | No | ||
| 63 | TNMD | 11273 | -0.781 | 0.1660 | No | ||
| 64 | EVX1 | 11569 | -0.859 | 0.1527 | No | ||
| 65 | SIN3A | 11586 | -0.863 | 0.1545 | No | ||
| 66 | RARG | 12337 | -1.060 | 0.1173 | No | ||
| 67 | SLC31A1 | 12832 | -1.210 | 0.0943 | No | ||
| 68 | NNAT | 12909 | -1.233 | 0.0940 | No | ||
| 69 | ADRB1 | 13116 | -1.295 | 0.0869 | No | ||
| 70 | PDCD1LG2 | 13248 | -1.337 | 0.0839 | No | ||
| 71 | IER5 | 13474 | -1.413 | 0.0761 | No | ||
| 72 | KLF12 | 13523 | -1.432 | 0.0780 | No | ||
| 73 | FOXP2 | 13796 | -1.525 | 0.0680 | No | ||
| 74 | MCC | 13818 | -1.536 | 0.0716 | No | ||
| 75 | TMOD3 | 14027 | -1.613 | 0.0653 | No | ||
| 76 | TECTA | 14542 | -1.831 | 0.0432 | No | ||
| 77 | PRDM1 | 14786 | -1.938 | 0.0361 | No | ||
| 78 | PHTF2 | 14988 | -2.040 | 0.0315 | No | ||
| 79 | ALDH1A2 | 15038 | -2.069 | 0.0352 | No | ||
| 80 | SHOX2 | 15065 | -2.080 | 0.0402 | No | ||
| 81 | HR | 15521 | -2.365 | 0.0230 | No | ||
| 82 | TBX4 | 15650 | -2.455 | 0.0236 | No | ||
| 83 | POU3F3 | 15720 | -2.510 | 0.0276 | No | ||
| 84 | AP1G2 | 15813 | -2.603 | 0.0307 | No | ||
| 85 | FERD3L | 15820 | -2.612 | 0.0384 | No | ||
| 86 | HSPG2 | 16442 | -3.223 | 0.0148 | No | ||
| 87 | PDE7A | 16602 | -3.454 | 0.0169 | No | ||
| 88 | TAF5L | 16671 | -3.560 | 0.0242 | No | ||
| 89 | HOXC4 | 16736 | -3.657 | 0.0320 | No | ||
| 90 | HEPH | 16779 | -3.734 | 0.0412 | No | ||
| 91 | NEDD4 | 17006 | -4.114 | 0.0417 | No | ||
| 92 | NCDN | 17016 | -4.130 | 0.0539 | No | ||
| 93 | SEMA5B | 17065 | -4.237 | 0.0644 | No | ||
| 94 | LMO4 | 17762 | -6.261 | 0.0461 | No |