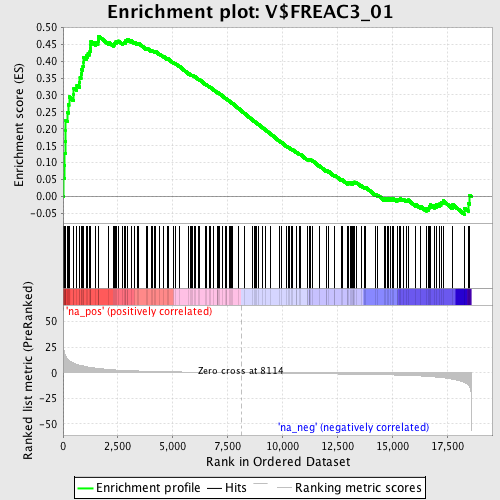
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FREAC3_01 |
| Enrichment Score (ES) | 0.47382483 |
| Normalized Enrichment Score (NES) | 1.6726419 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.055509914 |
| FWER p-Value | 0.544 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IMPDH1 | 16 | 28.243 | 0.0554 | Yes | ||
| 2 | TCF7L2 | 59 | 19.757 | 0.0925 | Yes | ||
| 3 | MMP11 | 81 | 18.609 | 0.1285 | Yes | ||
| 4 | CDKN1A | 100 | 17.229 | 0.1619 | Yes | ||
| 5 | LEF1 | 114 | 16.418 | 0.1939 | Yes | ||
| 6 | GFRA1 | 120 | 16.047 | 0.2256 | Yes | ||
| 7 | SQSTM1 | 189 | 13.689 | 0.2492 | Yes | ||
| 8 | GRB7 | 241 | 12.611 | 0.2716 | Yes | ||
| 9 | LAMB3 | 269 | 12.143 | 0.2944 | Yes | ||
| 10 | TIAM1 | 483 | 9.458 | 0.3017 | Yes | ||
| 11 | MBNL2 | 490 | 9.423 | 0.3201 | Yes | ||
| 12 | ZFP28 | 628 | 8.180 | 0.3290 | Yes | ||
| 13 | EFNA5 | 750 | 7.452 | 0.3373 | Yes | ||
| 14 | LCP2 | 768 | 7.342 | 0.3510 | Yes | ||
| 15 | BCL11A | 818 | 7.061 | 0.3624 | Yes | ||
| 16 | THRA | 838 | 6.988 | 0.3753 | Yes | ||
| 17 | PIP5K2A | 902 | 6.727 | 0.3853 | Yes | ||
| 18 | IDH1 | 927 | 6.636 | 0.3973 | Yes | ||
| 19 | HOXA9 | 932 | 6.609 | 0.4102 | Yes | ||
| 20 | TNFRSF19 | 1056 | 6.003 | 0.4155 | Yes | ||
| 21 | TGFB1 | 1133 | 5.662 | 0.4227 | Yes | ||
| 22 | CDKN1C | 1222 | 5.357 | 0.4286 | Yes | ||
| 23 | SUPT3H | 1247 | 5.284 | 0.4378 | Yes | ||
| 24 | PPP1R12A | 1258 | 5.250 | 0.4478 | Yes | ||
| 25 | SH3GL3 | 1266 | 5.222 | 0.4578 | Yes | ||
| 26 | DLGAP4 | 1481 | 4.635 | 0.4554 | Yes | ||
| 27 | HOXA4 | 1591 | 4.346 | 0.4582 | Yes | ||
| 28 | UTX | 1610 | 4.282 | 0.4658 | Yes | ||
| 29 | TCF15 | 1619 | 4.260 | 0.4738 | Yes | ||
| 30 | PAK1 | 2078 | 3.295 | 0.4556 | No | ||
| 31 | BCL9 | 2290 | 2.952 | 0.4500 | No | ||
| 32 | ETV1 | 2339 | 2.859 | 0.4531 | No | ||
| 33 | EYA1 | 2380 | 2.802 | 0.4565 | No | ||
| 34 | PELI2 | 2448 | 2.708 | 0.4583 | No | ||
| 35 | CDK6 | 2525 | 2.634 | 0.4594 | No | ||
| 36 | DCX | 2725 | 2.407 | 0.4535 | No | ||
| 37 | HOXC5 | 2779 | 2.353 | 0.4553 | No | ||
| 38 | CALD1 | 2832 | 2.299 | 0.4570 | No | ||
| 39 | POU2F1 | 2859 | 2.270 | 0.4602 | No | ||
| 40 | TBXAS1 | 2924 | 2.198 | 0.4611 | No | ||
| 41 | FGB | 2936 | 2.190 | 0.4648 | No | ||
| 42 | ITGB6 | 3094 | 2.060 | 0.4604 | No | ||
| 43 | TBC1D17 | 3241 | 1.944 | 0.4564 | No | ||
| 44 | LUC7L2 | 3374 | 1.832 | 0.4529 | No | ||
| 45 | TRIM8 | 3449 | 1.785 | 0.4525 | No | ||
| 46 | PHF15 | 3782 | 1.567 | 0.4376 | No | ||
| 47 | DVL2 | 3837 | 1.535 | 0.4377 | No | ||
| 48 | CNNM3 | 4014 | 1.432 | 0.4310 | No | ||
| 49 | TRPC4 | 4084 | 1.392 | 0.4301 | No | ||
| 50 | GNAO1 | 4157 | 1.353 | 0.4289 | No | ||
| 51 | MAP4K5 | 4210 | 1.327 | 0.4287 | No | ||
| 52 | MITF | 4405 | 1.227 | 0.4206 | No | ||
| 53 | HOXA11 | 4567 | 1.154 | 0.4142 | No | ||
| 54 | ULK1 | 4747 | 1.086 | 0.4067 | No | ||
| 55 | CD68 | 4781 | 1.068 | 0.4070 | No | ||
| 56 | CITED2 | 5036 | 0.967 | 0.3952 | No | ||
| 57 | ICAM5 | 5099 | 0.943 | 0.3937 | No | ||
| 58 | RAB3IP | 5300 | 0.866 | 0.3846 | No | ||
| 59 | EIF4EBP2 | 5736 | 0.702 | 0.3624 | No | ||
| 60 | SESTD1 | 5815 | 0.677 | 0.3595 | No | ||
| 61 | POGZ | 5873 | 0.657 | 0.3578 | No | ||
| 62 | HOXA3 | 5879 | 0.657 | 0.3588 | No | ||
| 63 | ANGPT2 | 5975 | 0.629 | 0.3549 | No | ||
| 64 | SLC38A3 | 6011 | 0.616 | 0.3542 | No | ||
| 65 | SCG2 | 6171 | 0.572 | 0.3468 | No | ||
| 66 | FGF9 | 6230 | 0.552 | 0.3447 | No | ||
| 67 | CHN1 | 6480 | 0.474 | 0.3322 | No | ||
| 68 | MYL1 | 6552 | 0.452 | 0.3292 | No | ||
| 69 | BAMBI | 6651 | 0.428 | 0.3248 | No | ||
| 70 | NOG | 6696 | 0.416 | 0.3232 | No | ||
| 71 | ZIC5 | 6873 | 0.363 | 0.3144 | No | ||
| 72 | CPNE1 | 7032 | 0.314 | 0.3065 | No | ||
| 73 | SIM1 | 7049 | 0.309 | 0.3062 | No | ||
| 74 | GCG | 7050 | 0.309 | 0.3068 | No | ||
| 75 | SCRN3 | 7082 | 0.301 | 0.3057 | No | ||
| 76 | LHX5 | 7113 | 0.293 | 0.3047 | No | ||
| 77 | NEUROG1 | 7251 | 0.253 | 0.2978 | No | ||
| 78 | BRUNOL6 | 7388 | 0.212 | 0.2908 | No | ||
| 79 | HOXA2 | 7432 | 0.198 | 0.2889 | No | ||
| 80 | NTN1 | 7460 | 0.190 | 0.2878 | No | ||
| 81 | SPDEF | 7583 | 0.149 | 0.2815 | No | ||
| 82 | ZIC2 | 7632 | 0.137 | 0.2792 | No | ||
| 83 | PCF11 | 7692 | 0.121 | 0.2762 | No | ||
| 84 | LHX9 | 7703 | 0.116 | 0.2759 | No | ||
| 85 | BUB3 | 8006 | 0.028 | 0.2596 | No | ||
| 86 | MRGPRF | 8273 | -0.047 | 0.2453 | No | ||
| 87 | PURA | 8623 | -0.136 | 0.2266 | No | ||
| 88 | KCTD4 | 8737 | -0.165 | 0.2208 | No | ||
| 89 | NEUROD2 | 8773 | -0.174 | 0.2193 | No | ||
| 90 | ESRRG | 8830 | -0.188 | 0.2166 | No | ||
| 91 | ETV4 | 8890 | -0.201 | 0.2138 | No | ||
| 92 | BRMS1 | 9104 | -0.252 | 0.2028 | No | ||
| 93 | SLC35A2 | 9201 | -0.277 | 0.1981 | No | ||
| 94 | HOXA10 | 9464 | -0.340 | 0.1846 | No | ||
| 95 | PPP1CB | 9879 | -0.446 | 0.1631 | No | ||
| 96 | GSH1 | 9953 | -0.464 | 0.1600 | No | ||
| 97 | TGFB3 | 10184 | -0.524 | 0.1486 | No | ||
| 98 | PHOX2B | 10261 | -0.547 | 0.1456 | No | ||
| 99 | LRAT | 10310 | -0.558 | 0.1441 | No | ||
| 100 | OTX2 | 10429 | -0.588 | 0.1389 | No | ||
| 101 | VAX1 | 10464 | -0.595 | 0.1382 | No | ||
| 102 | OTP | 10614 | -0.631 | 0.1314 | No | ||
| 103 | EFEMP1 | 10655 | -0.640 | 0.1305 | No | ||
| 104 | RKHD3 | 10790 | -0.670 | 0.1246 | No | ||
| 105 | LGI1 | 10823 | -0.678 | 0.1242 | No | ||
| 106 | ATOH8 | 11123 | -0.744 | 0.1095 | No | ||
| 107 | PAPPA | 11156 | -0.755 | 0.1093 | No | ||
| 108 | PLUNC | 11213 | -0.767 | 0.1077 | No | ||
| 109 | LRP2 | 11218 | -0.768 | 0.1091 | No | ||
| 110 | EGR2 | 11225 | -0.769 | 0.1103 | No | ||
| 111 | PCSK1 | 11275 | -0.781 | 0.1092 | No | ||
| 112 | SLITRK6 | 11372 | -0.806 | 0.1056 | No | ||
| 113 | ATP5G2 | 11669 | -0.882 | 0.0913 | No | ||
| 114 | PPIL5 | 11988 | -0.967 | 0.0760 | No | ||
| 115 | C1QTNF6 | 12024 | -0.977 | 0.0760 | No | ||
| 116 | ARID4A | 12109 | -1.000 | 0.0735 | No | ||
| 117 | AGPAT4 | 12371 | -1.070 | 0.0615 | No | ||
| 118 | HERPUD1 | 12375 | -1.070 | 0.0634 | No | ||
| 119 | FOSL2 | 12675 | -1.163 | 0.0496 | No | ||
| 120 | MAFF | 12717 | -1.175 | 0.0497 | No | ||
| 121 | FBXO40 | 12969 | -1.251 | 0.0386 | No | ||
| 122 | TCERG1L | 12976 | -1.255 | 0.0407 | No | ||
| 123 | ZHX2 | 13005 | -1.264 | 0.0417 | No | ||
| 124 | HOXA13 | 13095 | -1.289 | 0.0395 | No | ||
| 125 | TTR | 13132 | -1.301 | 0.0401 | No | ||
| 126 | NRG1 | 13205 | -1.321 | 0.0389 | No | ||
| 127 | HOXC12 | 13220 | -1.328 | 0.0408 | No | ||
| 128 | BCL11B | 13246 | -1.337 | 0.0421 | No | ||
| 129 | GRIK3 | 13280 | -1.348 | 0.0430 | No | ||
| 130 | BNC2 | 13350 | -1.373 | 0.0420 | No | ||
| 131 | DTNA | 13594 | -1.457 | 0.0317 | No | ||
| 132 | SDK2 | 13751 | -1.506 | 0.0262 | No | ||
| 133 | FOXP2 | 13796 | -1.525 | 0.0269 | No | ||
| 134 | DCN | 14232 | -1.691 | 0.0067 | No | ||
| 135 | GPR1 | 14339 | -1.735 | 0.0044 | No | ||
| 136 | PRICKLE2 | 14657 | -1.882 | -0.0090 | No | ||
| 137 | FAP | 14671 | -1.890 | -0.0059 | No | ||
| 138 | PRDM1 | 14786 | -1.938 | -0.0083 | No | ||
| 139 | AKT2 | 14825 | -1.957 | -0.0064 | No | ||
| 140 | A2BP1 | 14906 | -2.002 | -0.0068 | No | ||
| 141 | TRERF1 | 14999 | -2.046 | -0.0077 | No | ||
| 142 | TIMM9 | 15058 | -2.078 | -0.0067 | No | ||
| 143 | HOXC8 | 15219 | -2.170 | -0.0110 | No | ||
| 144 | ARTN | 15242 | -2.181 | -0.0079 | No | ||
| 145 | MRVI1 | 15326 | -2.234 | -0.0079 | No | ||
| 146 | AKT1S1 | 15371 | -2.265 | -0.0058 | No | ||
| 147 | TIAL1 | 15496 | -2.348 | -0.0078 | No | ||
| 148 | TBX4 | 15650 | -2.455 | -0.0112 | No | ||
| 149 | POU3F3 | 15720 | -2.510 | -0.0099 | No | ||
| 150 | RASSF6 | 16073 | -2.844 | -0.0234 | No | ||
| 151 | NTRK3 | 16280 | -3.049 | -0.0284 | No | ||
| 152 | MCM7 | 16549 | -3.369 | -0.0362 | No | ||
| 153 | BRUNOL4 | 16642 | -3.515 | -0.0342 | No | ||
| 154 | AP4M1 | 16680 | -3.570 | -0.0291 | No | ||
| 155 | TRAF3 | 16727 | -3.648 | -0.0243 | No | ||
| 156 | PPP1R10 | 16912 | -3.938 | -0.0264 | No | ||
| 157 | NEDD4 | 17006 | -4.114 | -0.0233 | No | ||
| 158 | XPO7 | 17144 | -4.405 | -0.0219 | No | ||
| 159 | PRPF39 | 17224 | -4.594 | -0.0171 | No | ||
| 160 | MPP6 | 17316 | -4.802 | -0.0124 | No | ||
| 161 | LMO4 | 17762 | -6.261 | -0.0240 | No | ||
| 162 | FSTL1 | 18298 | -9.456 | -0.0342 | No | ||
| 163 | EMP3 | 18487 | -12.090 | -0.0203 | No | ||
| 164 | MRPS18B | 18534 | -13.647 | 0.0044 | No |