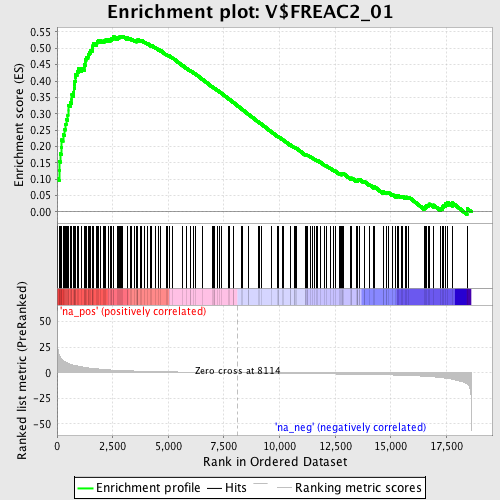
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$FREAC2_01 |
| Enrichment Score (ES) | 0.53725755 |
| Normalized Enrichment Score (NES) | 1.912657 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015915012 |
| FWER p-Value | 0.023 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 60.375 | 0.1011 | Yes | ||
| 2 | PRKCH | 92 | 17.715 | 0.1258 | Yes | ||
| 3 | CDKN1A | 100 | 17.229 | 0.1543 | Yes | ||
| 4 | MARCKS | 141 | 15.219 | 0.1776 | Yes | ||
| 5 | SQSTM1 | 189 | 13.689 | 0.1980 | Yes | ||
| 6 | NDRG1 | 200 | 13.450 | 0.2200 | Yes | ||
| 7 | SMAD1 | 266 | 12.210 | 0.2369 | Yes | ||
| 8 | HBP1 | 346 | 10.961 | 0.2509 | Yes | ||
| 9 | SEPP1 | 361 | 10.781 | 0.2682 | Yes | ||
| 10 | ERG | 405 | 10.291 | 0.2831 | Yes | ||
| 11 | TIAM1 | 483 | 9.458 | 0.2948 | Yes | ||
| 12 | MBNL2 | 490 | 9.423 | 0.3103 | Yes | ||
| 13 | ABTB1 | 491 | 9.420 | 0.3260 | Yes | ||
| 14 | MMP13 | 588 | 8.467 | 0.3350 | Yes | ||
| 15 | RAB6IP1 | 658 | 7.944 | 0.3446 | Yes | ||
| 16 | HOXB4 | 664 | 7.921 | 0.3576 | Yes | ||
| 17 | KLF3 | 743 | 7.475 | 0.3658 | Yes | ||
| 18 | NFIX | 759 | 7.390 | 0.3774 | Yes | ||
| 19 | ETV5 | 784 | 7.256 | 0.3883 | Yes | ||
| 20 | DUSP1 | 797 | 7.185 | 0.3996 | Yes | ||
| 21 | BCL11A | 818 | 7.061 | 0.4104 | Yes | ||
| 22 | THRA | 838 | 6.988 | 0.4210 | Yes | ||
| 23 | LRP5 | 897 | 6.757 | 0.4292 | Yes | ||
| 24 | SMAD5 | 952 | 6.520 | 0.4372 | Yes | ||
| 25 | PPAP2B | 1091 | 5.848 | 0.4395 | Yes | ||
| 26 | CDKN1C | 1222 | 5.357 | 0.4414 | Yes | ||
| 27 | IGF1 | 1226 | 5.346 | 0.4502 | Yes | ||
| 28 | AXL | 1263 | 5.231 | 0.4570 | Yes | ||
| 29 | SH3GL3 | 1266 | 5.222 | 0.4657 | Yes | ||
| 30 | MBNL1 | 1300 | 5.110 | 0.4724 | Yes | ||
| 31 | TBCC | 1389 | 4.867 | 0.4758 | Yes | ||
| 32 | UHRF2 | 1420 | 4.791 | 0.4822 | Yes | ||
| 33 | NAP1L3 | 1463 | 4.685 | 0.4878 | Yes | ||
| 34 | BCAS3 | 1509 | 4.553 | 0.4929 | Yes | ||
| 35 | KBTBD2 | 1575 | 4.401 | 0.4968 | Yes | ||
| 36 | STK3 | 1581 | 4.367 | 0.5038 | Yes | ||
| 37 | UTX | 1610 | 4.282 | 0.5095 | Yes | ||
| 38 | TCF15 | 1619 | 4.260 | 0.5162 | Yes | ||
| 39 | EGLN1 | 1779 | 3.904 | 0.5141 | Yes | ||
| 40 | PPM1D | 1793 | 3.877 | 0.5199 | Yes | ||
| 41 | CHD2 | 1845 | 3.767 | 0.5234 | Yes | ||
| 42 | HIBADH | 1964 | 3.507 | 0.5229 | Yes | ||
| 43 | PAK1 | 2078 | 3.295 | 0.5223 | Yes | ||
| 44 | DHRS3 | 2137 | 3.202 | 0.5245 | Yes | ||
| 45 | ZADH2 | 2195 | 3.119 | 0.5266 | Yes | ||
| 46 | BCL9 | 2290 | 2.952 | 0.5265 | Yes | ||
| 47 | EYA1 | 2380 | 2.802 | 0.5263 | Yes | ||
| 48 | PELI2 | 2448 | 2.708 | 0.5272 | Yes | ||
| 49 | HEBP2 | 2455 | 2.701 | 0.5314 | Yes | ||
| 50 | EXT1 | 2517 | 2.642 | 0.5326 | Yes | ||
| 51 | ELMO1 | 2524 | 2.634 | 0.5366 | Yes | ||
| 52 | VPS52 | 2724 | 2.409 | 0.5299 | Yes | ||
| 53 | HIPK1 | 2744 | 2.390 | 0.5329 | Yes | ||
| 54 | DUSP4 | 2783 | 2.350 | 0.5347 | Yes | ||
| 55 | CALD1 | 2832 | 2.299 | 0.5360 | Yes | ||
| 56 | BRIP1 | 2913 | 2.212 | 0.5353 | Yes | ||
| 57 | DNAJB12 | 2946 | 2.180 | 0.5373 | Yes | ||
| 58 | RBM4 | 3145 | 2.017 | 0.5299 | No | ||
| 59 | NGEF | 3159 | 2.008 | 0.5326 | No | ||
| 60 | IRS4 | 3283 | 1.913 | 0.5291 | No | ||
| 61 | PITPNC1 | 3359 | 1.846 | 0.5281 | No | ||
| 62 | UBE2H | 3495 | 1.747 | 0.5237 | No | ||
| 63 | IL6ST | 3579 | 1.698 | 0.5221 | No | ||
| 64 | RASD1 | 3627 | 1.669 | 0.5223 | No | ||
| 65 | TCF1 | 3628 | 1.668 | 0.5251 | No | ||
| 66 | NRAP | 3631 | 1.667 | 0.5278 | No | ||
| 67 | PLAG1 | 3769 | 1.574 | 0.5230 | No | ||
| 68 | PHF15 | 3782 | 1.567 | 0.5250 | No | ||
| 69 | FOXB1 | 3943 | 1.470 | 0.5187 | No | ||
| 70 | TRPC4 | 4084 | 1.392 | 0.5135 | No | ||
| 71 | MAP4K5 | 4210 | 1.327 | 0.5089 | No | ||
| 72 | PITX2 | 4241 | 1.312 | 0.5095 | No | ||
| 73 | MITF | 4405 | 1.227 | 0.5027 | No | ||
| 74 | HOXA11 | 4567 | 1.154 | 0.4959 | No | ||
| 75 | RORA | 4661 | 1.116 | 0.4927 | No | ||
| 76 | ROR1 | 4908 | 1.019 | 0.4811 | No | ||
| 77 | VIT | 4980 | 0.986 | 0.4789 | No | ||
| 78 | CITED2 | 5036 | 0.967 | 0.4775 | No | ||
| 79 | STOML3 | 5202 | 0.907 | 0.4701 | No | ||
| 80 | BMP5 | 5649 | 0.731 | 0.4471 | No | ||
| 81 | FGF12 | 5820 | 0.676 | 0.4390 | No | ||
| 82 | CD109 | 6006 | 0.617 | 0.4300 | No | ||
| 83 | SLC38A3 | 6011 | 0.616 | 0.4309 | No | ||
| 84 | KCTD15 | 6140 | 0.583 | 0.4249 | No | ||
| 85 | FGF9 | 6230 | 0.552 | 0.4210 | No | ||
| 86 | SYT1 | 6540 | 0.456 | 0.4050 | No | ||
| 87 | GREM1 | 6978 | 0.328 | 0.3818 | No | ||
| 88 | CPNE1 | 7032 | 0.314 | 0.3795 | No | ||
| 89 | HHIP | 7066 | 0.305 | 0.3782 | No | ||
| 90 | SCRN3 | 7082 | 0.301 | 0.3779 | No | ||
| 91 | PLCB2 | 7228 | 0.261 | 0.3705 | No | ||
| 92 | GNPNAT1 | 7283 | 0.243 | 0.3680 | No | ||
| 93 | BRUNOL6 | 7388 | 0.212 | 0.3627 | No | ||
| 94 | PCF11 | 7692 | 0.121 | 0.3464 | No | ||
| 95 | PIK3C2A | 7753 | 0.100 | 0.3434 | No | ||
| 96 | NFIB | 7907 | 0.060 | 0.3352 | No | ||
| 97 | HOXD9 | 7932 | 0.052 | 0.3339 | No | ||
| 98 | MRGPRF | 8273 | -0.047 | 0.3156 | No | ||
| 99 | CHCHD7 | 8323 | -0.061 | 0.3130 | No | ||
| 100 | RAD21 | 8607 | -0.132 | 0.2979 | No | ||
| 101 | PURA | 8623 | -0.136 | 0.2973 | No | ||
| 102 | RBM16 | 9050 | -0.241 | 0.2746 | No | ||
| 103 | HAND2 | 9106 | -0.253 | 0.2721 | No | ||
| 104 | WEE1 | 9178 | -0.271 | 0.2687 | No | ||
| 105 | EPHB3 | 9634 | -0.379 | 0.2446 | No | ||
| 106 | EZH1 | 9888 | -0.450 | 0.2316 | No | ||
| 107 | ADAM12 | 9939 | -0.462 | 0.2297 | No | ||
| 108 | FBXW7 | 9957 | -0.465 | 0.2296 | No | ||
| 109 | RASGEF1B | 10113 | -0.507 | 0.2220 | No | ||
| 110 | TGFB3 | 10184 | -0.524 | 0.2191 | No | ||
| 111 | SLITRK5 | 10468 | -0.595 | 0.2047 | No | ||
| 112 | OFCC1 | 10473 | -0.596 | 0.2055 | No | ||
| 113 | CGA | 10659 | -0.641 | 0.1966 | No | ||
| 114 | ENPP2 | 10681 | -0.645 | 0.1965 | No | ||
| 115 | NNT | 10722 | -0.652 | 0.1954 | No | ||
| 116 | PROX1 | 10777 | -0.669 | 0.1936 | No | ||
| 117 | PAPPA | 11156 | -0.755 | 0.1744 | No | ||
| 118 | BMP2 | 11180 | -0.760 | 0.1744 | No | ||
| 119 | PLUNC | 11213 | -0.767 | 0.1739 | No | ||
| 120 | SLC34A3 | 11261 | -0.777 | 0.1727 | No | ||
| 121 | PCSK1 | 11275 | -0.781 | 0.1733 | No | ||
| 122 | EN2 | 11375 | -0.807 | 0.1693 | No | ||
| 123 | NFRKB | 11498 | -0.841 | 0.1641 | No | ||
| 124 | SIN3A | 11586 | -0.863 | 0.1608 | No | ||
| 125 | CXXC5 | 11661 | -0.881 | 0.1583 | No | ||
| 126 | DSTN | 11724 | -0.897 | 0.1564 | No | ||
| 127 | RARA | 11825 | -0.924 | 0.1525 | No | ||
| 128 | GPRC5C | 12030 | -0.980 | 0.1431 | No | ||
| 129 | ARID4A | 12109 | -1.000 | 0.1405 | No | ||
| 130 | MLLT6 | 12275 | -1.044 | 0.1333 | No | ||
| 131 | EVI1 | 12416 | -1.086 | 0.1276 | No | ||
| 132 | GPR4 | 12497 | -1.108 | 0.1251 | No | ||
| 133 | FOSL2 | 12675 | -1.163 | 0.1174 | No | ||
| 134 | MAFF | 12717 | -1.175 | 0.1172 | No | ||
| 135 | PCDH18 | 12801 | -1.198 | 0.1147 | No | ||
| 136 | BDNF | 12829 | -1.208 | 0.1152 | No | ||
| 137 | CD2AP | 12837 | -1.211 | 0.1169 | No | ||
| 138 | DHX8 | 12854 | -1.215 | 0.1181 | No | ||
| 139 | NRG1 | 13205 | -1.321 | 0.1013 | No | ||
| 140 | HOXC12 | 13220 | -1.328 | 0.1028 | No | ||
| 141 | BCL11B | 13246 | -1.337 | 0.1036 | No | ||
| 142 | HABP4 | 13434 | -1.399 | 0.0958 | No | ||
| 143 | CDX2 | 13481 | -1.416 | 0.0957 | No | ||
| 144 | ACCN1 | 13522 | -1.431 | 0.0959 | No | ||
| 145 | KLF12 | 13523 | -1.432 | 0.0983 | No | ||
| 146 | SLC2A4 | 13569 | -1.447 | 0.0983 | No | ||
| 147 | DTNA | 13594 | -1.457 | 0.0995 | No | ||
| 148 | FOXP2 | 13796 | -1.525 | 0.0911 | No | ||
| 149 | MCC | 13818 | -1.536 | 0.0925 | No | ||
| 150 | CLPX | 14055 | -1.622 | 0.0825 | No | ||
| 151 | FGF20 | 14237 | -1.693 | 0.0755 | No | ||
| 152 | POU3F4 | 14260 | -1.702 | 0.0771 | No | ||
| 153 | SYT6 | 14656 | -1.881 | 0.0589 | No | ||
| 154 | TPP2 | 14658 | -1.882 | 0.0620 | No | ||
| 155 | PRDM1 | 14786 | -1.938 | 0.0583 | No | ||
| 156 | AKT2 | 14825 | -1.957 | 0.0595 | No | ||
| 157 | MID1 | 14907 | -2.003 | 0.0585 | No | ||
| 158 | CLDN8 | 15072 | -2.082 | 0.0531 | No | ||
| 159 | HOXC8 | 15219 | -2.170 | 0.0488 | No | ||
| 160 | EN1 | 15285 | -2.206 | 0.0490 | No | ||
| 161 | MRVI1 | 15326 | -2.234 | 0.0505 | No | ||
| 162 | ATBF1 | 15464 | -2.327 | 0.0470 | No | ||
| 163 | HR | 15521 | -2.365 | 0.0479 | No | ||
| 164 | TBX4 | 15650 | -2.455 | 0.0451 | No | ||
| 165 | POU3F3 | 15720 | -2.510 | 0.0456 | No | ||
| 166 | AP1G2 | 15813 | -2.603 | 0.0449 | No | ||
| 167 | E2F3 | 16497 | -3.306 | 0.0134 | No | ||
| 168 | MCM7 | 16549 | -3.369 | 0.0163 | No | ||
| 169 | PDE7A | 16602 | -3.454 | 0.0193 | No | ||
| 170 | AP4M1 | 16680 | -3.570 | 0.0211 | No | ||
| 171 | HOXC4 | 16736 | -3.657 | 0.0242 | No | ||
| 172 | RPS18 | 16916 | -3.948 | 0.0211 | No | ||
| 173 | STOML2 | 17231 | -4.613 | 0.0118 | No | ||
| 174 | MPP6 | 17316 | -4.802 | 0.0153 | No | ||
| 175 | DSG4 | 17377 | -4.982 | 0.0204 | No | ||
| 176 | TOB1 | 17459 | -5.211 | 0.0247 | No | ||
| 177 | DOCK3 | 17549 | -5.495 | 0.0291 | No | ||
| 178 | LMO4 | 17762 | -6.261 | 0.0281 | No | ||
| 179 | CPN1 | 18427 | -10.870 | 0.0103 | No |