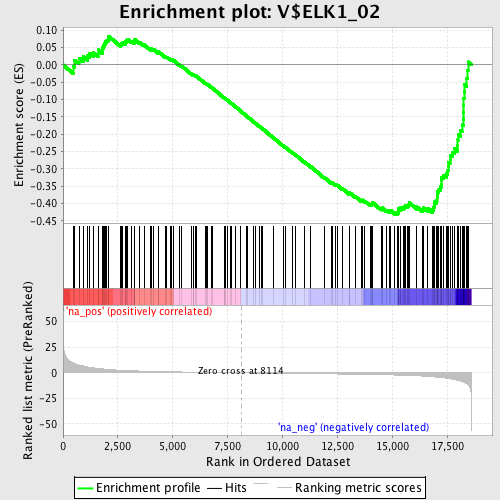
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$ELK1_02 |
| Enrichment Score (ES) | -0.4319229 |
| Normalized Enrichment Score (NES) | -1.5928065 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.08544766 |
| FWER p-Value | 0.721 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ITM2C | 460 | 9.734 | -0.0031 | No | ||
| 2 | PRKACB | 519 | 9.130 | 0.0143 | No | ||
| 3 | KRTCAP2 | 726 | 7.549 | 0.0201 | No | ||
| 4 | POLL | 921 | 6.659 | 0.0245 | No | ||
| 5 | RAB11B | 1126 | 5.712 | 0.0263 | No | ||
| 6 | DPP8 | 1208 | 5.401 | 0.0340 | No | ||
| 7 | TBCC | 1389 | 4.867 | 0.0352 | No | ||
| 8 | RAP2C | 1594 | 4.342 | 0.0339 | No | ||
| 9 | FBXW9 | 1595 | 4.330 | 0.0436 | No | ||
| 10 | INSIG2 | 1797 | 3.862 | 0.0414 | No | ||
| 11 | SEC24C | 1802 | 3.855 | 0.0498 | No | ||
| 12 | HMG20A | 1827 | 3.802 | 0.0570 | No | ||
| 13 | FBXO3 | 1874 | 3.703 | 0.0629 | No | ||
| 14 | NEDD1 | 1924 | 3.591 | 0.0683 | No | ||
| 15 | BCL2L1 | 1998 | 3.459 | 0.0721 | No | ||
| 16 | TBX6 | 2054 | 3.349 | 0.0766 | No | ||
| 17 | CGGBP1 | 2069 | 3.318 | 0.0833 | No | ||
| 18 | AP1B1 | 2607 | 2.543 | 0.0599 | No | ||
| 19 | UQCRH | 2674 | 2.461 | 0.0619 | No | ||
| 20 | VPS52 | 2724 | 2.409 | 0.0646 | No | ||
| 21 | PHKB | 2828 | 2.301 | 0.0642 | No | ||
| 22 | FBXO36 | 2852 | 2.279 | 0.0681 | No | ||
| 23 | STK4 | 2902 | 2.227 | 0.0704 | No | ||
| 24 | MAPKAP1 | 2911 | 2.212 | 0.0750 | No | ||
| 25 | ARHGAP1 | 3104 | 2.051 | 0.0692 | No | ||
| 26 | TBC1D17 | 3241 | 1.944 | 0.0662 | No | ||
| 27 | NR1H2 | 3259 | 1.931 | 0.0696 | No | ||
| 28 | PEX11A | 3265 | 1.929 | 0.0736 | No | ||
| 29 | PCYT1A | 3482 | 1.764 | 0.0659 | No | ||
| 30 | CRB3 | 3691 | 1.627 | 0.0583 | No | ||
| 31 | TRO | 4004 | 1.435 | 0.0446 | No | ||
| 32 | NF1 | 4013 | 1.433 | 0.0474 | No | ||
| 33 | SYT5 | 4098 | 1.383 | 0.0459 | No | ||
| 34 | PURB | 4327 | 1.262 | 0.0364 | No | ||
| 35 | ITGB1BP1 | 4347 | 1.248 | 0.0382 | No | ||
| 36 | PCQAP | 4659 | 1.116 | 0.0239 | No | ||
| 37 | RAPH1 | 4725 | 1.095 | 0.0228 | No | ||
| 38 | GOLGA1 | 4884 | 1.026 | 0.0166 | No | ||
| 39 | MYST2 | 4940 | 1.006 | 0.0158 | No | ||
| 40 | RPL19 | 5040 | 0.966 | 0.0126 | No | ||
| 41 | MAPK8IP3 | 5293 | 0.869 | 0.0009 | No | ||
| 42 | ZNF22 | 5413 | 0.825 | -0.0036 | No | ||
| 43 | ZBTB11 | 5851 | 0.665 | -0.0258 | No | ||
| 44 | ELAVL1 | 5945 | 0.637 | -0.0294 | No | ||
| 45 | SHC1 | 5949 | 0.636 | -0.0282 | No | ||
| 46 | NUDT12 | 6019 | 0.614 | -0.0305 | No | ||
| 47 | SIRT6 | 6070 | 0.604 | -0.0319 | No | ||
| 48 | PPIE | 6467 | 0.480 | -0.0522 | No | ||
| 49 | UBR1 | 6546 | 0.454 | -0.0554 | No | ||
| 50 | ZDHHC5 | 6581 | 0.446 | -0.0563 | No | ||
| 51 | TRIM46 | 6766 | 0.392 | -0.0654 | No | ||
| 52 | BAT3 | 6827 | 0.373 | -0.0678 | No | ||
| 53 | FOXH1 | 7341 | 0.227 | -0.0950 | No | ||
| 54 | LSM5 | 7362 | 0.221 | -0.0956 | No | ||
| 55 | STX12 | 7415 | 0.204 | -0.0980 | No | ||
| 56 | GGTL3 | 7514 | 0.171 | -0.1029 | No | ||
| 57 | GTF3C2 | 7650 | 0.132 | -0.1099 | No | ||
| 58 | ACP2 | 7666 | 0.128 | -0.1104 | No | ||
| 59 | COG3 | 7871 | 0.069 | -0.1213 | No | ||
| 60 | FBXO22 | 8071 | 0.011 | -0.1321 | No | ||
| 61 | COX8A | 8350 | -0.070 | -0.1470 | No | ||
| 62 | NFYA | 8419 | -0.085 | -0.1505 | No | ||
| 63 | DDOST | 8665 | -0.147 | -0.1634 | No | ||
| 64 | CLDN7 | 8780 | -0.176 | -0.1692 | No | ||
| 65 | LYPLA2 | 8934 | -0.212 | -0.1770 | No | ||
| 66 | IL2RG | 9022 | -0.235 | -0.1812 | No | ||
| 67 | ACTR2 | 9109 | -0.254 | -0.1853 | No | ||
| 68 | DIABLO | 9585 | -0.369 | -0.2102 | No | ||
| 69 | MTX2 | 10046 | -0.490 | -0.2340 | No | ||
| 70 | SUPT5H | 10128 | -0.510 | -0.2372 | No | ||
| 71 | UFD1L | 10456 | -0.593 | -0.2536 | No | ||
| 72 | CLDN5 | 10577 | -0.622 | -0.2587 | No | ||
| 73 | CPSF3 | 10994 | -0.714 | -0.2796 | No | ||
| 74 | UBE2L3 | 11286 | -0.783 | -0.2936 | No | ||
| 75 | HMGA1 | 11904 | -0.945 | -0.3249 | No | ||
| 76 | NDUFS1 | 12247 | -1.037 | -0.3411 | No | ||
| 77 | TSTA3 | 12261 | -1.041 | -0.3394 | No | ||
| 78 | RBM22 | 12410 | -1.083 | -0.3450 | No | ||
| 79 | FMR1 | 12490 | -1.106 | -0.3468 | No | ||
| 80 | OGG1 | 12730 | -1.180 | -0.3571 | No | ||
| 81 | LMAN2 | 13041 | -1.275 | -0.3710 | No | ||
| 82 | GTF2A2 | 13058 | -1.279 | -0.3690 | No | ||
| 83 | LEPRE1 | 13311 | -1.358 | -0.3796 | No | ||
| 84 | GTF3C1 | 13603 | -1.460 | -0.3921 | No | ||
| 85 | PSMC6 | 13626 | -1.466 | -0.3900 | No | ||
| 86 | PSMC1 | 13752 | -1.507 | -0.3934 | No | ||
| 87 | NKIRAS2 | 14002 | -1.605 | -0.4033 | No | ||
| 88 | TRIM44 | 14061 | -1.624 | -0.4028 | No | ||
| 89 | MARK3 | 14089 | -1.637 | -0.4006 | No | ||
| 90 | BMP2K | 14118 | -1.648 | -0.3984 | No | ||
| 91 | TRAF7 | 14499 | -1.810 | -0.4149 | No | ||
| 92 | GABARAPL2 | 14537 | -1.829 | -0.4128 | No | ||
| 93 | UBE2N | 14737 | -1.915 | -0.4193 | No | ||
| 94 | AP1M1 | 14891 | -1.995 | -0.4231 | No | ||
| 95 | DHX30 | 14921 | -2.008 | -0.4202 | No | ||
| 96 | SON | 15122 | -2.116 | -0.4262 | No | ||
| 97 | NAT5 | 15228 | -2.173 | -0.4270 | Yes | ||
| 98 | CALU | 15269 | -2.197 | -0.4243 | Yes | ||
| 99 | KLHDC3 | 15280 | -2.204 | -0.4199 | Yes | ||
| 100 | EN1 | 15285 | -2.206 | -0.4151 | Yes | ||
| 101 | AKT1S1 | 15371 | -2.265 | -0.4147 | Yes | ||
| 102 | ZNF23 | 15387 | -2.273 | -0.4104 | Yes | ||
| 103 | SMUG1 | 15526 | -2.370 | -0.4125 | Yes | ||
| 104 | ARCN1 | 15576 | -2.399 | -0.4098 | Yes | ||
| 105 | MTPN | 15603 | -2.421 | -0.4058 | Yes | ||
| 106 | SAMD8 | 15718 | -2.506 | -0.4063 | Yes | ||
| 107 | EPN1 | 15759 | -2.550 | -0.4028 | Yes | ||
| 108 | HSPC171 | 15778 | -2.565 | -0.3980 | Yes | ||
| 109 | PSMC2 | 16121 | -2.892 | -0.4100 | Yes | ||
| 110 | RPL37 | 16392 | -3.152 | -0.4176 | Yes | ||
| 111 | ATP5A1 | 16439 | -3.220 | -0.4128 | Yes | ||
| 112 | PSMD12 | 16629 | -3.498 | -0.4152 | Yes | ||
| 113 | CRK | 16826 | -3.808 | -0.4173 | Yes | ||
| 114 | DDX5 | 16866 | -3.864 | -0.4107 | Yes | ||
| 115 | HAT1 | 16908 | -3.934 | -0.4041 | Yes | ||
| 116 | RPS18 | 16916 | -3.948 | -0.3956 | Yes | ||
| 117 | NRAS | 17023 | -4.143 | -0.3921 | Yes | ||
| 118 | CHERP | 17041 | -4.190 | -0.3836 | Yes | ||
| 119 | TIMM8B | 17052 | -4.213 | -0.3747 | Yes | ||
| 120 | TCF7 | 17067 | -4.240 | -0.3659 | Yes | ||
| 121 | SDHD | 17130 | -4.383 | -0.3595 | Yes | ||
| 122 | SRP19 | 17182 | -4.503 | -0.3521 | Yes | ||
| 123 | EIF4G1 | 17226 | -4.598 | -0.3441 | Yes | ||
| 124 | STOML2 | 17231 | -4.613 | -0.3340 | Yes | ||
| 125 | MRPL13 | 17248 | -4.651 | -0.3244 | Yes | ||
| 126 | MTBP | 17338 | -4.870 | -0.3183 | Yes | ||
| 127 | DNAJC7 | 17488 | -5.299 | -0.3145 | Yes | ||
| 128 | MIR16 | 17537 | -5.464 | -0.3048 | Yes | ||
| 129 | TOMM40 | 17562 | -5.530 | -0.2937 | Yes | ||
| 130 | UBADC1 | 17572 | -5.558 | -0.2817 | Yes | ||
| 131 | CDC45L | 17635 | -5.759 | -0.2722 | Yes | ||
| 132 | EEF1B2 | 17658 | -5.823 | -0.2603 | Yes | ||
| 133 | DNAJA2 | 17761 | -6.250 | -0.2518 | Yes | ||
| 134 | SLC30A7 | 17827 | -6.552 | -0.2406 | Yes | ||
| 135 | TUFM | 17991 | -7.329 | -0.2330 | Yes | ||
| 136 | IMMT | 17995 | -7.360 | -0.2166 | Yes | ||
| 137 | RFC4 | 18000 | -7.383 | -0.2003 | Yes | ||
| 138 | PRKAG2 | 18108 | -7.981 | -0.1882 | Yes | ||
| 139 | EIF5A | 18184 | -8.462 | -0.1733 | Yes | ||
| 140 | FANCD2 | 18230 | -8.839 | -0.1559 | Yes | ||
| 141 | GART | 18243 | -8.950 | -0.1364 | Yes | ||
| 142 | EXTL2 | 18255 | -9.040 | -0.1167 | Yes | ||
| 143 | SNRPB | 18260 | -9.079 | -0.0966 | Yes | ||
| 144 | IPO4 | 18281 | -9.238 | -0.0769 | Yes | ||
| 145 | SMS | 18305 | -9.549 | -0.0567 | Yes | ||
| 146 | MRPL3 | 18403 | -10.562 | -0.0383 | Yes | ||
| 147 | TRMT1 | 18420 | -10.801 | -0.0149 | Yes | ||
| 148 | PAPD4 | 18457 | -11.360 | 0.0086 | Yes |