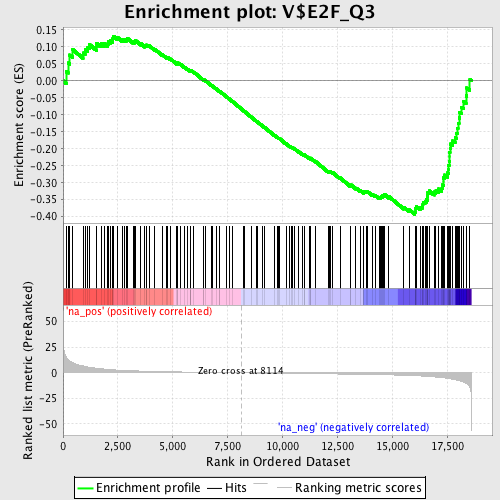
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | V$E2F_Q3 |
| Enrichment Score (ES) | -0.3934702 |
| Normalized Enrichment Score (NES) | -1.4646822 |
| Nominal p-value | 0.005376344 |
| FDR q-value | 0.12719809 |
| FWER p-Value | 0.984 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | MXD3 | 143 | 15.133 | 0.0275 | No | ||
| 2 | DCK | 229 | 12.823 | 0.0527 | No | ||
| 3 | LASP1 | 301 | 11.682 | 0.0761 | No | ||
| 4 | MEIS1 | 440 | 9.904 | 0.0917 | No | ||
| 5 | DMD | 908 | 6.710 | 0.0820 | No | ||
| 6 | SSBP3 | 1003 | 6.269 | 0.0915 | No | ||
| 7 | RAB11B | 1126 | 5.712 | 0.0982 | No | ||
| 8 | CTCF | 1205 | 5.422 | 0.1066 | No | ||
| 9 | PIAS1 | 1518 | 4.536 | 0.1003 | No | ||
| 10 | DPYSL2 | 1527 | 4.522 | 0.1104 | No | ||
| 11 | EZH2 | 1726 | 4.027 | 0.1091 | No | ||
| 12 | PAPOLG | 1873 | 3.703 | 0.1098 | No | ||
| 13 | ATE1 | 2040 | 3.376 | 0.1086 | No | ||
| 14 | TBX6 | 2054 | 3.349 | 0.1157 | No | ||
| 15 | EPHB2 | 2148 | 3.187 | 0.1181 | No | ||
| 16 | GCH1 | 2251 | 3.033 | 0.1197 | No | ||
| 17 | CTDSP1 | 2267 | 3.007 | 0.1258 | No | ||
| 18 | ZCCHC8 | 2289 | 2.953 | 0.1316 | No | ||
| 19 | PIM1 | 2474 | 2.686 | 0.1279 | No | ||
| 20 | HIST1H2BK | 2684 | 2.447 | 0.1223 | No | ||
| 21 | H2AFZ | 2795 | 2.336 | 0.1217 | No | ||
| 22 | KLF5 | 2885 | 2.244 | 0.1221 | No | ||
| 23 | HIST1H2AH | 2934 | 2.192 | 0.1247 | No | ||
| 24 | DDB2 | 3185 | 1.990 | 0.1157 | No | ||
| 25 | CACNA1G | 3273 | 1.924 | 0.1155 | No | ||
| 26 | PHF13 | 3309 | 1.889 | 0.1180 | No | ||
| 27 | ERBB2IP | 3546 | 1.714 | 0.1092 | No | ||
| 28 | CBX5 | 3730 | 1.605 | 0.1031 | No | ||
| 29 | PHF15 | 3782 | 1.567 | 0.1040 | No | ||
| 30 | ARHGAP6 | 3820 | 1.542 | 0.1055 | No | ||
| 31 | NUP153 | 3923 | 1.488 | 0.1035 | No | ||
| 32 | KLF4 | 4178 | 1.344 | 0.0929 | No | ||
| 33 | ONECUT1 | 4509 | 1.179 | 0.0777 | No | ||
| 34 | PEG3 | 4730 | 1.092 | 0.0684 | No | ||
| 35 | AP1S2 | 4767 | 1.075 | 0.0689 | No | ||
| 36 | HMGN2 | 4895 | 1.023 | 0.0644 | No | ||
| 37 | TREX2 | 5185 | 0.912 | 0.0509 | No | ||
| 38 | PHF12 | 5198 | 0.909 | 0.0524 | No | ||
| 39 | ADAMTS2 | 5209 | 0.905 | 0.0539 | No | ||
| 40 | HS6ST3 | 5345 | 0.849 | 0.0486 | No | ||
| 41 | CASP8AP2 | 5544 | 0.772 | 0.0397 | No | ||
| 42 | OLFML3 | 5689 | 0.716 | 0.0335 | No | ||
| 43 | SNPH | 5784 | 0.689 | 0.0301 | No | ||
| 44 | UXT | 5793 | 0.684 | 0.0312 | No | ||
| 45 | RHD | 5822 | 0.674 | 0.0313 | No | ||
| 46 | NOL4 | 5956 | 0.634 | 0.0256 | No | ||
| 47 | HNRPD | 6399 | 0.500 | 0.0028 | No | ||
| 48 | APRIN | 6402 | 0.499 | 0.0038 | No | ||
| 49 | EPHB1 | 6490 | 0.470 | 0.0002 | No | ||
| 50 | STK35 | 6751 | 0.396 | -0.0129 | No | ||
| 51 | BAT3 | 6827 | 0.373 | -0.0161 | No | ||
| 52 | HIST1H1D | 6976 | 0.329 | -0.0234 | No | ||
| 53 | LHX5 | 7113 | 0.293 | -0.0301 | No | ||
| 54 | PLK4 | 7144 | 0.283 | -0.0310 | No | ||
| 55 | NDUFA11 | 7431 | 0.199 | -0.0460 | No | ||
| 56 | FMO4 | 7578 | 0.151 | -0.0536 | No | ||
| 57 | E2F7 | 7698 | 0.118 | -0.0598 | No | ||
| 58 | PODN | 8203 | -0.026 | -0.0870 | No | ||
| 59 | GRIK2 | 8266 | -0.044 | -0.0902 | No | ||
| 60 | TBX3 | 8605 | -0.132 | -0.1082 | No | ||
| 61 | FLI1 | 8812 | -0.183 | -0.1190 | No | ||
| 62 | CPNE5 | 8850 | -0.192 | -0.1205 | No | ||
| 63 | SLC38A1 | 9094 | -0.251 | -0.1331 | No | ||
| 64 | WEE1 | 9178 | -0.271 | -0.1370 | No | ||
| 65 | STAG2 | 9638 | -0.380 | -0.1609 | No | ||
| 66 | SEZ6 | 9767 | -0.416 | -0.1669 | No | ||
| 67 | TOPBP1 | 9813 | -0.426 | -0.1683 | No | ||
| 68 | USP1 | 9852 | -0.437 | -0.1694 | No | ||
| 69 | PAQR4 | 10171 | -0.520 | -0.1854 | No | ||
| 70 | BARHL1 | 10325 | -0.564 | -0.1923 | No | ||
| 71 | PCNA | 10397 | -0.580 | -0.1948 | No | ||
| 72 | UFD1L | 10456 | -0.593 | -0.1966 | No | ||
| 73 | STMN1 | 10556 | -0.615 | -0.2005 | No | ||
| 74 | RBBP4 | 10733 | -0.655 | -0.2085 | No | ||
| 75 | SLC25A14 | 10898 | -0.696 | -0.2158 | No | ||
| 76 | NPR3 | 10996 | -0.714 | -0.2194 | No | ||
| 77 | ATRX | 10997 | -0.715 | -0.2177 | No | ||
| 78 | SFRS1 | 11226 | -0.769 | -0.2283 | No | ||
| 79 | MCM8 | 11271 | -0.780 | -0.2288 | No | ||
| 80 | PCSK1 | 11275 | -0.781 | -0.2272 | No | ||
| 81 | ADCY8 | 11499 | -0.842 | -0.2373 | No | ||
| 82 | ARID4A | 12109 | -1.000 | -0.2679 | No | ||
| 83 | ASCL1 | 12162 | -1.016 | -0.2684 | No | ||
| 84 | CLTA | 12176 | -1.019 | -0.2667 | No | ||
| 85 | POLE2 | 12276 | -1.045 | -0.2696 | No | ||
| 86 | MAZ | 12622 | -1.147 | -0.2856 | No | ||
| 87 | HNRPA1 | 13113 | -1.295 | -0.3092 | No | ||
| 88 | FBXO5 | 13120 | -1.297 | -0.3065 | No | ||
| 89 | CNOT3 | 13346 | -1.372 | -0.3155 | No | ||
| 90 | TRIM47 | 13552 | -1.443 | -0.3232 | No | ||
| 91 | SLC25A3 | 13705 | -1.491 | -0.3280 | No | ||
| 92 | SYT11 | 13707 | -1.491 | -0.3245 | No | ||
| 93 | SEMA5A | 13805 | -1.529 | -0.3262 | No | ||
| 94 | KCNA1 | 13877 | -1.561 | -0.3264 | No | ||
| 95 | NRP2 | 14078 | -1.631 | -0.3335 | No | ||
| 96 | DNMT1 | 14222 | -1.686 | -0.3373 | No | ||
| 97 | KCND2 | 14409 | -1.766 | -0.3432 | No | ||
| 98 | SLC6A4 | 14481 | -1.802 | -0.3429 | No | ||
| 99 | JPH1 | 14509 | -1.814 | -0.3401 | No | ||
| 100 | RFC1 | 14556 | -1.839 | -0.3383 | No | ||
| 101 | MCM6 | 14599 | -1.854 | -0.3363 | No | ||
| 102 | SALL1 | 14650 | -1.878 | -0.3346 | No | ||
| 103 | PRPS1 | 14849 | -1.970 | -0.3408 | No | ||
| 104 | IPO7 | 15529 | -2.373 | -0.3720 | No | ||
| 105 | PRKDC | 15786 | -2.569 | -0.3799 | No | ||
| 106 | MDGA1 | 16038 | -2.817 | -0.3869 | Yes | ||
| 107 | LIG1 | 16050 | -2.827 | -0.3809 | Yes | ||
| 108 | CDCA7 | 16052 | -2.829 | -0.3744 | Yes | ||
| 109 | PVRL1 | 16089 | -2.862 | -0.3697 | Yes | ||
| 110 | INSM1 | 16276 | -3.048 | -0.3726 | Yes | ||
| 111 | DLST | 16363 | -3.128 | -0.3700 | Yes | ||
| 112 | TFRC | 16396 | -3.157 | -0.3644 | Yes | ||
| 113 | MCM2 | 16404 | -3.166 | -0.3574 | Yes | ||
| 114 | E2F3 | 16497 | -3.306 | -0.3547 | Yes | ||
| 115 | MCM7 | 16549 | -3.369 | -0.3496 | Yes | ||
| 116 | ATAD2 | 16605 | -3.457 | -0.3445 | Yes | ||
| 117 | DNAJC11 | 16611 | -3.468 | -0.3367 | Yes | ||
| 118 | MCM4 | 16623 | -3.480 | -0.3292 | Yes | ||
| 119 | AP4M1 | 16680 | -3.570 | -0.3239 | Yes | ||
| 120 | MCM3 | 16907 | -3.932 | -0.3270 | Yes | ||
| 121 | PPP1CC | 16994 | -4.092 | -0.3222 | Yes | ||
| 122 | GMNN | 17110 | -4.344 | -0.3183 | Yes | ||
| 123 | PCSK4 | 17263 | -4.683 | -0.3156 | Yes | ||
| 124 | NCL | 17298 | -4.766 | -0.3063 | Yes | ||
| 125 | SMAD6 | 17333 | -4.861 | -0.2969 | Yes | ||
| 126 | PPP1R8 | 17334 | -4.861 | -0.2855 | Yes | ||
| 127 | CDC25A | 17396 | -5.014 | -0.2772 | Yes | ||
| 128 | HTF9C | 17515 | -5.363 | -0.2711 | Yes | ||
| 129 | CDC6 | 17565 | -5.540 | -0.2608 | Yes | ||
| 130 | ING3 | 17574 | -5.561 | -0.2483 | Yes | ||
| 131 | RBBP7 | 17593 | -5.614 | -0.2362 | Yes | ||
| 132 | SFRS2 | 17611 | -5.660 | -0.2240 | Yes | ||
| 133 | E2F1 | 17613 | -5.669 | -0.2108 | Yes | ||
| 134 | CDC45L | 17635 | -5.759 | -0.1985 | Yes | ||
| 135 | NASP | 17651 | -5.801 | -0.1858 | Yes | ||
| 136 | RAD51 | 17755 | -6.221 | -0.1769 | Yes | ||
| 137 | ORC1L | 17868 | -6.700 | -0.1674 | Yes | ||
| 138 | KPNB1 | 17917 | -6.958 | -0.1538 | Yes | ||
| 139 | POLA2 | 17962 | -7.207 | -0.1394 | Yes | ||
| 140 | MAT2A | 18001 | -7.392 | -0.1243 | Yes | ||
| 141 | UNG | 18046 | -7.689 | -0.1087 | Yes | ||
| 142 | MRPL18 | 18076 | -7.848 | -0.0920 | Yes | ||
| 143 | TCP1 | 18163 | -8.325 | -0.0773 | Yes | ||
| 144 | RANBP1 | 18261 | -9.082 | -0.0614 | Yes | ||
| 145 | IMPDH2 | 18367 | -10.029 | -0.0437 | Yes | ||
| 146 | ABCF2 | 18400 | -10.546 | -0.0209 | Yes | ||
| 147 | SHMT1 | 18542 | -13.985 | 0.0040 | Yes |