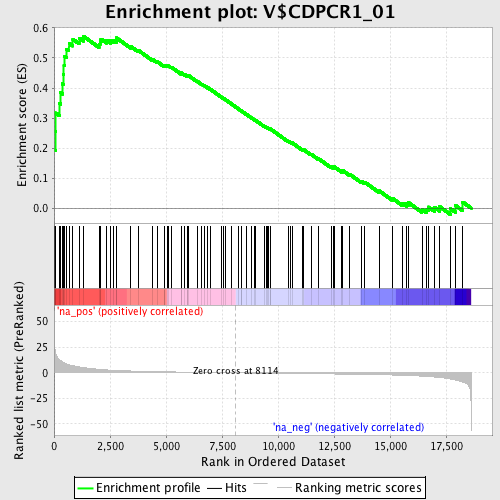
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$CDPCR1_01 |
| Enrichment Score (ES) | 0.5728243 |
| Normalized Enrichment Score (NES) | 1.8303167 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.02818061 |
| FWER p-Value | 0.097 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | EMP1 | 0 | 60.375 | 0.1954 | Yes | ||
| 2 | TCF7L2 | 59 | 19.757 | 0.2563 | Yes | ||
| 3 | IL15 | 74 | 18.963 | 0.3169 | Yes | ||
| 4 | GRB7 | 241 | 12.611 | 0.3488 | Yes | ||
| 5 | PPT2 | 292 | 11.788 | 0.3842 | Yes | ||
| 6 | BTBD3 | 378 | 10.592 | 0.4139 | Yes | ||
| 7 | ERG | 405 | 10.291 | 0.4458 | Yes | ||
| 8 | MEIS1 | 440 | 9.904 | 0.4761 | Yes | ||
| 9 | TLE4 | 473 | 9.560 | 0.5053 | Yes | ||
| 10 | EGR1 | 550 | 8.793 | 0.5296 | Yes | ||
| 11 | HOXB4 | 664 | 7.921 | 0.5492 | Yes | ||
| 12 | THRA | 838 | 6.988 | 0.5625 | Yes | ||
| 13 | FCHSD1 | 1118 | 5.758 | 0.5661 | Yes | ||
| 14 | MBNL1 | 1300 | 5.110 | 0.5728 | Yes | ||
| 15 | CREM | 2007 | 3.448 | 0.5459 | No | ||
| 16 | TBX6 | 2054 | 3.349 | 0.5542 | No | ||
| 17 | LYPLA3 | 2087 | 3.285 | 0.5632 | No | ||
| 18 | ETV1 | 2339 | 2.859 | 0.5589 | No | ||
| 19 | EXT1 | 2517 | 2.642 | 0.5579 | No | ||
| 20 | DIO2 | 2633 | 2.513 | 0.5598 | No | ||
| 21 | HOXC5 | 2779 | 2.353 | 0.5596 | No | ||
| 22 | DUSP4 | 2783 | 2.350 | 0.5670 | No | ||
| 23 | COL2A1 | 3416 | 1.804 | 0.5388 | No | ||
| 24 | PAX8 | 3752 | 1.590 | 0.5258 | No | ||
| 25 | NR3C2 | 4369 | 1.239 | 0.4966 | No | ||
| 26 | ST7 | 4601 | 1.144 | 0.4878 | No | ||
| 27 | NTNG2 | 4909 | 1.019 | 0.4746 | No | ||
| 28 | MYST2 | 4940 | 1.006 | 0.4762 | No | ||
| 29 | CUL2 | 5047 | 0.963 | 0.4736 | No | ||
| 30 | HOXC6 | 5103 | 0.941 | 0.4737 | No | ||
| 31 | PDCD10 | 5227 | 0.896 | 0.4699 | No | ||
| 32 | STAT5B | 5666 | 0.724 | 0.4486 | No | ||
| 33 | LIN28 | 5677 | 0.720 | 0.4504 | No | ||
| 34 | FGF12 | 5820 | 0.676 | 0.4450 | No | ||
| 35 | INSRR | 5939 | 0.638 | 0.4407 | No | ||
| 36 | NOL4 | 5956 | 0.634 | 0.4419 | No | ||
| 37 | MAOB | 5995 | 0.623 | 0.4418 | No | ||
| 38 | ITGA7 | 6400 | 0.500 | 0.4216 | No | ||
| 39 | DGKB | 6596 | 0.442 | 0.4125 | No | ||
| 40 | NOG | 6696 | 0.416 | 0.4085 | No | ||
| 41 | NKX2-2 | 6861 | 0.365 | 0.4009 | No | ||
| 42 | HOXD10 | 6977 | 0.329 | 0.3957 | No | ||
| 43 | SERPING1 | 7484 | 0.182 | 0.3690 | No | ||
| 44 | ADAM10 | 7548 | 0.163 | 0.3661 | No | ||
| 45 | STMN2 | 7648 | 0.132 | 0.3612 | No | ||
| 46 | NFIB | 7907 | 0.060 | 0.3475 | No | ||
| 47 | KCND1 | 8212 | -0.027 | 0.3312 | No | ||
| 48 | AP3M2 | 8365 | -0.073 | 0.3232 | No | ||
| 49 | GRIK4 | 8375 | -0.075 | 0.3230 | No | ||
| 50 | TBX3 | 8605 | -0.132 | 0.3110 | No | ||
| 51 | NHLH2 | 8798 | -0.181 | 0.3013 | No | ||
| 52 | RPH3A | 8946 | -0.215 | 0.2940 | No | ||
| 53 | PLXNA3 | 8984 | -0.225 | 0.2928 | No | ||
| 54 | RFX4 | 9375 | -0.320 | 0.2727 | No | ||
| 55 | HOXA10 | 9464 | -0.340 | 0.2691 | No | ||
| 56 | CPNE6 | 9516 | -0.350 | 0.2675 | No | ||
| 57 | DPP10 | 9589 | -0.370 | 0.2648 | No | ||
| 58 | STAG2 | 9638 | -0.380 | 0.2634 | No | ||
| 59 | PIGN | 9653 | -0.383 | 0.2639 | No | ||
| 60 | VAX1 | 10464 | -0.595 | 0.2221 | No | ||
| 61 | NTRK1 | 10545 | -0.614 | 0.2198 | No | ||
| 62 | KCNS1 | 10622 | -0.632 | 0.2177 | No | ||
| 63 | SLC12A5 | 11093 | -0.737 | 0.1948 | No | ||
| 64 | HOXB2 | 11151 | -0.753 | 0.1941 | No | ||
| 65 | DBC1 | 11469 | -0.834 | 0.1797 | No | ||
| 66 | MGAT5B | 11781 | -0.911 | 0.1659 | No | ||
| 67 | NRXN3 | 12368 | -1.068 | 0.1377 | No | ||
| 68 | EEF1A2 | 12480 | -1.104 | 0.1353 | No | ||
| 69 | CYP26B1 | 12504 | -1.109 | 0.1376 | No | ||
| 70 | SERPINI1 | 12845 | -1.213 | 0.1232 | No | ||
| 71 | RTN4RL2 | 12867 | -1.221 | 0.1260 | No | ||
| 72 | INSM2 | 13196 | -1.318 | 0.1126 | No | ||
| 73 | MAG | 13715 | -1.494 | 0.0895 | No | ||
| 74 | KCNA1 | 13877 | -1.561 | 0.0859 | No | ||
| 75 | SFTPC | 14513 | -1.816 | 0.0575 | No | ||
| 76 | SON | 15122 | -2.116 | 0.0315 | No | ||
| 77 | KLF15 | 15559 | -2.390 | 0.0157 | No | ||
| 78 | PFN2 | 15730 | -2.518 | 0.0147 | No | ||
| 79 | CSMD3 | 15819 | -2.609 | 0.0184 | No | ||
| 80 | TRIB2 | 16422 | -3.191 | -0.0038 | No | ||
| 81 | BRUNOL4 | 16642 | -3.515 | -0.0042 | No | ||
| 82 | BHLHB3 | 16712 | -3.613 | 0.0038 | No | ||
| 83 | CIB3 | 16976 | -4.051 | 0.0027 | No | ||
| 84 | GATA4 | 17214 | -4.569 | 0.0047 | No | ||
| 85 | NR2F6 | 17695 | -5.981 | -0.0019 | No | ||
| 86 | KDELR2 | 17913 | -6.947 | 0.0089 | No | ||
| 87 | GART | 18243 | -8.950 | 0.0201 | No |