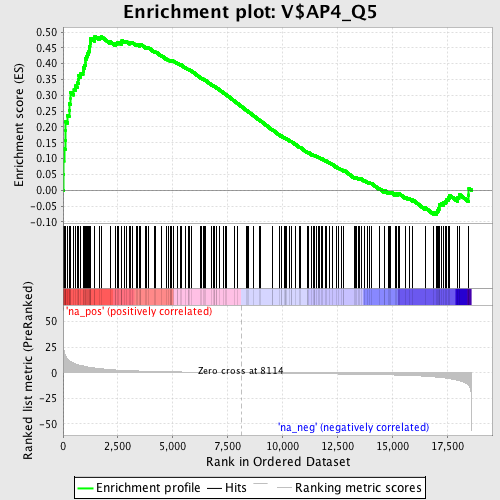
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | V$AP4_Q5 |
| Enrichment Score (ES) | 0.48706064 |
| Normalized Enrichment Score (NES) | 1.7267376 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05305609 |
| FWER p-Value | 0.337 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IMPDH1 | 16 | 28.243 | 0.0489 | Yes | ||
| 2 | ABR | 25 | 25.496 | 0.0933 | Yes | ||
| 3 | PPP1R9B | 41 | 21.764 | 0.1308 | Yes | ||
| 4 | CUL7 | 96 | 17.443 | 0.1586 | Yes | ||
| 5 | SYTL1 | 104 | 16.860 | 0.1879 | Yes | ||
| 6 | ESR1 | 111 | 16.546 | 0.2168 | Yes | ||
| 7 | PRKCQ | 196 | 13.515 | 0.2360 | Yes | ||
| 8 | LZTS2 | 275 | 12.026 | 0.2529 | Yes | ||
| 9 | RUNX1 | 286 | 11.889 | 0.2733 | Yes | ||
| 10 | BACH1 | 342 | 11.060 | 0.2898 | Yes | ||
| 11 | HBP1 | 346 | 10.961 | 0.3090 | Yes | ||
| 12 | MESDC1 | 495 | 9.369 | 0.3175 | Yes | ||
| 13 | RAB27A | 554 | 8.772 | 0.3298 | Yes | ||
| 14 | MAP3K3 | 642 | 8.089 | 0.3393 | Yes | ||
| 15 | TRAF4 | 700 | 7.724 | 0.3498 | Yes | ||
| 16 | PCYT1B | 710 | 7.648 | 0.3628 | Yes | ||
| 17 | SLC25A24 | 808 | 7.121 | 0.3701 | Yes | ||
| 18 | DMD | 908 | 6.710 | 0.3765 | Yes | ||
| 19 | MOSPD1 | 919 | 6.676 | 0.3877 | Yes | ||
| 20 | STIM1 | 986 | 6.327 | 0.3953 | Yes | ||
| 21 | CACNB2 | 1012 | 6.221 | 0.4049 | Yes | ||
| 22 | FMO5 | 1016 | 6.193 | 0.4156 | Yes | ||
| 23 | TNFRSF19 | 1056 | 6.003 | 0.4241 | Yes | ||
| 24 | RIT1 | 1112 | 5.776 | 0.4313 | Yes | ||
| 25 | PGF | 1172 | 5.511 | 0.4378 | Yes | ||
| 26 | HSD11B1 | 1190 | 5.453 | 0.4465 | Yes | ||
| 27 | ZRANB1 | 1211 | 5.390 | 0.4549 | Yes | ||
| 28 | IGF1 | 1226 | 5.346 | 0.4635 | Yes | ||
| 29 | MBP | 1234 | 5.321 | 0.4725 | Yes | ||
| 30 | CASKIN2 | 1262 | 5.235 | 0.4803 | Yes | ||
| 31 | NDST2 | 1430 | 4.751 | 0.4796 | Yes | ||
| 32 | VAMP3 | 1446 | 4.715 | 0.4871 | Yes | ||
| 33 | DDIT4 | 1664 | 4.149 | 0.4826 | No | ||
| 34 | ABHD4 | 1743 | 3.970 | 0.4854 | No | ||
| 35 | DHRS3 | 2137 | 3.202 | 0.4697 | No | ||
| 36 | EYA1 | 2380 | 2.802 | 0.4615 | No | ||
| 37 | RAI1 | 2404 | 2.768 | 0.4651 | No | ||
| 38 | CD79B | 2464 | 2.692 | 0.4667 | No | ||
| 39 | WNT10B | 2544 | 2.606 | 0.4670 | No | ||
| 40 | HOXB5 | 2652 | 2.484 | 0.4656 | No | ||
| 41 | APBA1 | 2669 | 2.465 | 0.4690 | No | ||
| 42 | VAT1 | 2680 | 2.449 | 0.4728 | No | ||
| 43 | SCN4A | 2784 | 2.347 | 0.4713 | No | ||
| 44 | HIP1 | 2869 | 2.265 | 0.4708 | No | ||
| 45 | EPOR | 3032 | 2.109 | 0.4657 | No | ||
| 46 | ZNF385 | 3067 | 2.081 | 0.4675 | No | ||
| 47 | PTK7 | 3155 | 2.010 | 0.4664 | No | ||
| 48 | SLC7A8 | 3342 | 1.859 | 0.4595 | No | ||
| 49 | ATP1B4 | 3399 | 1.816 | 0.4597 | No | ||
| 50 | UBE2H | 3495 | 1.747 | 0.4576 | No | ||
| 51 | SUV39H2 | 3519 | 1.730 | 0.4594 | No | ||
| 52 | XPNPEP1 | 3534 | 1.720 | 0.4617 | No | ||
| 53 | ARHGEF2 | 3773 | 1.571 | 0.4516 | No | ||
| 54 | IL11 | 3819 | 1.542 | 0.4518 | No | ||
| 55 | RXRG | 3905 | 1.496 | 0.4499 | No | ||
| 56 | CHD4 | 4159 | 1.353 | 0.4385 | No | ||
| 57 | TRAM1 | 4232 | 1.317 | 0.4369 | No | ||
| 58 | RGS7 | 4500 | 1.184 | 0.4246 | No | ||
| 59 | RBMS3 | 4713 | 1.097 | 0.4150 | No | ||
| 60 | PDGFB | 4810 | 1.057 | 0.4117 | No | ||
| 61 | HMGN2 | 4895 | 1.023 | 0.4089 | No | ||
| 62 | BAT5 | 4916 | 1.015 | 0.4096 | No | ||
| 63 | FZD9 | 4917 | 1.015 | 0.4114 | No | ||
| 64 | MYO1C | 4951 | 1.002 | 0.4114 | No | ||
| 65 | CITED2 | 5036 | 0.967 | 0.4085 | No | ||
| 66 | DNAJA1 | 5203 | 0.906 | 0.4011 | No | ||
| 67 | UBE2D3 | 5234 | 0.895 | 0.4011 | No | ||
| 68 | FOXD3 | 5329 | 0.855 | 0.3975 | No | ||
| 69 | MAP3K13 | 5377 | 0.837 | 0.3964 | No | ||
| 70 | KCNE1L | 5572 | 0.759 | 0.3872 | No | ||
| 71 | ADRBK1 | 5693 | 0.715 | 0.3820 | No | ||
| 72 | HSD3B7 | 5717 | 0.709 | 0.3820 | No | ||
| 73 | KCNH2 | 5765 | 0.693 | 0.3806 | No | ||
| 74 | ATRNL1 | 5871 | 0.658 | 0.3761 | No | ||
| 75 | MYF5 | 6272 | 0.538 | 0.3554 | No | ||
| 76 | SLC8A3 | 6323 | 0.523 | 0.3536 | No | ||
| 77 | SMARCA1 | 6375 | 0.510 | 0.3517 | No | ||
| 78 | DOCK4 | 6432 | 0.492 | 0.3495 | No | ||
| 79 | BAP1 | 6475 | 0.477 | 0.3481 | No | ||
| 80 | HOXA6 | 6755 | 0.395 | 0.3337 | No | ||
| 81 | RTKN | 6833 | 0.372 | 0.3301 | No | ||
| 82 | SOST | 6894 | 0.357 | 0.3275 | No | ||
| 83 | PDLIM4 | 6900 | 0.355 | 0.3279 | No | ||
| 84 | HPCA | 6990 | 0.326 | 0.3236 | No | ||
| 85 | CHRD | 7004 | 0.323 | 0.3235 | No | ||
| 86 | ANXA8 | 7117 | 0.290 | 0.3179 | No | ||
| 87 | SRGAP1 | 7318 | 0.232 | 0.3075 | No | ||
| 88 | MLLT7 | 7397 | 0.209 | 0.3036 | No | ||
| 89 | ASB5 | 7449 | 0.193 | 0.3012 | No | ||
| 90 | VDP | 7820 | 0.084 | 0.2813 | No | ||
| 91 | DLL4 | 7945 | 0.048 | 0.2747 | No | ||
| 92 | PCBP4 | 8351 | -0.070 | 0.2528 | No | ||
| 93 | ELF4 | 8385 | -0.077 | 0.2512 | No | ||
| 94 | DES | 8466 | -0.096 | 0.2470 | No | ||
| 95 | PRDM8 | 8689 | -0.153 | 0.2352 | No | ||
| 96 | PTCH2 | 8955 | -0.218 | 0.2212 | No | ||
| 97 | PDE3A | 8985 | -0.225 | 0.2201 | No | ||
| 98 | WNT4 | 9553 | -0.361 | 0.1900 | No | ||
| 99 | GPM6B | 9565 | -0.363 | 0.1900 | No | ||
| 100 | ERBB3 | 9870 | -0.443 | 0.1743 | No | ||
| 101 | ITGB1BP2 | 9932 | -0.460 | 0.1718 | No | ||
| 102 | KCNMA1 | 9945 | -0.463 | 0.1720 | No | ||
| 103 | HFE2 | 10068 | -0.496 | 0.1662 | No | ||
| 104 | ALDH1A1 | 10135 | -0.511 | 0.1636 | No | ||
| 105 | DLL3 | 10139 | -0.512 | 0.1643 | No | ||
| 106 | TGFB3 | 10184 | -0.524 | 0.1628 | No | ||
| 107 | CAPZA3 | 10333 | -0.566 | 0.1558 | No | ||
| 108 | DMC1 | 10392 | -0.579 | 0.1537 | No | ||
| 109 | WNT9A | 10401 | -0.580 | 0.1543 | No | ||
| 110 | AMOT | 10582 | -0.623 | 0.1456 | No | ||
| 111 | INSR | 10783 | -0.669 | 0.1359 | No | ||
| 112 | MID2 | 10804 | -0.673 | 0.1360 | No | ||
| 113 | ATOH8 | 11123 | -0.744 | 0.1201 | No | ||
| 114 | SYTL2 | 11137 | -0.749 | 0.1207 | No | ||
| 115 | SOX12 | 11198 | -0.764 | 0.1188 | No | ||
| 116 | NDST4 | 11339 | -0.797 | 0.1126 | No | ||
| 117 | HAPLN2 | 11397 | -0.813 | 0.1110 | No | ||
| 118 | SEMA6D | 11446 | -0.828 | 0.1098 | No | ||
| 119 | DBC1 | 11469 | -0.834 | 0.1101 | No | ||
| 120 | KCNJ2 | 11550 | -0.854 | 0.1073 | No | ||
| 121 | GPR21 | 11633 | -0.876 | 0.1044 | No | ||
| 122 | ASB16 | 11667 | -0.882 | 0.1041 | No | ||
| 123 | DEPDC2 | 11797 | -0.915 | 0.0987 | No | ||
| 124 | PHF7 | 11817 | -0.921 | 0.0993 | No | ||
| 125 | AMPH | 11977 | -0.964 | 0.0924 | No | ||
| 126 | GRIN2D | 12001 | -0.969 | 0.0929 | No | ||
| 127 | MASP1 | 12134 | -1.008 | 0.0875 | No | ||
| 128 | DAAM1 | 12271 | -1.043 | 0.0820 | No | ||
| 129 | PABPC5 | 12455 | -1.097 | 0.0740 | No | ||
| 130 | CPEB3 | 12560 | -1.126 | 0.0703 | No | ||
| 131 | FOSL2 | 12675 | -1.163 | 0.0662 | No | ||
| 132 | SLC12A8 | 12784 | -1.193 | 0.0624 | No | ||
| 133 | TNRC4 | 12793 | -1.195 | 0.0641 | No | ||
| 134 | GRIK3 | 13280 | -1.348 | 0.0401 | No | ||
| 135 | MYBPH | 13320 | -1.361 | 0.0404 | No | ||
| 136 | BNC2 | 13350 | -1.373 | 0.0412 | No | ||
| 137 | GNB3 | 13467 | -1.411 | 0.0374 | No | ||
| 138 | SULT2B1 | 13497 | -1.424 | 0.0384 | No | ||
| 139 | BTG1 | 13585 | -1.454 | 0.0362 | No | ||
| 140 | KCNN2 | 13716 | -1.494 | 0.0318 | No | ||
| 141 | CHRNG | 13875 | -1.561 | 0.0260 | No | ||
| 142 | IL17B | 13952 | -1.587 | 0.0247 | No | ||
| 143 | WNT2B | 14038 | -1.616 | 0.0229 | No | ||
| 144 | PSIP1 | 14412 | -1.767 | 0.0058 | No | ||
| 145 | UNC13B | 14638 | -1.872 | -0.0031 | No | ||
| 146 | SALL1 | 14650 | -1.878 | -0.0004 | No | ||
| 147 | AKT2 | 14825 | -1.957 | -0.0064 | No | ||
| 148 | OBSCN | 14875 | -1.988 | -0.0056 | No | ||
| 149 | C7ORF16 | 14940 | -2.014 | -0.0055 | No | ||
| 150 | PAICS | 15163 | -2.142 | -0.0137 | No | ||
| 151 | NR4A3 | 15188 | -2.149 | -0.0113 | No | ||
| 152 | AQP2 | 15264 | -2.195 | -0.0115 | No | ||
| 153 | TGFBR2 | 15324 | -2.233 | -0.0107 | No | ||
| 154 | TRAPPC3 | 15605 | -2.422 | -0.0216 | No | ||
| 155 | ANP32A | 15767 | -2.556 | -0.0259 | No | ||
| 156 | PRICKLE1 | 15931 | -2.714 | -0.0299 | No | ||
| 157 | RASGRF2 | 16506 | -3.317 | -0.0552 | No | ||
| 158 | PADI2 | 16897 | -3.918 | -0.0695 | No | ||
| 159 | NCDN | 17016 | -4.130 | -0.0686 | No | ||
| 160 | TCF7 | 17067 | -4.240 | -0.0638 | No | ||
| 161 | ENO3 | 17103 | -4.313 | -0.0581 | No | ||
| 162 | KLF13 | 17138 | -4.398 | -0.0522 | No | ||
| 163 | PTPRS | 17142 | -4.402 | -0.0446 | No | ||
| 164 | STOML2 | 17231 | -4.613 | -0.0413 | No | ||
| 165 | QTRTD1 | 17345 | -4.885 | -0.0388 | No | ||
| 166 | RPS6KB1 | 17420 | -5.068 | -0.0339 | No | ||
| 167 | TPM3 | 17486 | -5.295 | -0.0281 | No | ||
| 168 | RHOBTB2 | 17556 | -5.512 | -0.0221 | No | ||
| 169 | E2F1 | 17613 | -5.669 | -0.0152 | No | ||
| 170 | ARHGAP8 | 17988 | -7.307 | -0.0226 | No | ||
| 171 | ARHGAP24 | 18044 | -7.665 | -0.0121 | No | ||
| 172 | EMP3 | 18487 | -12.090 | -0.0148 | No | ||
| 173 | PPAT | 18495 | -12.319 | 0.0066 | No |