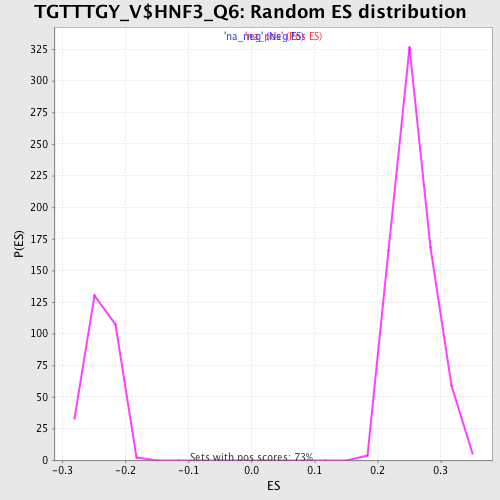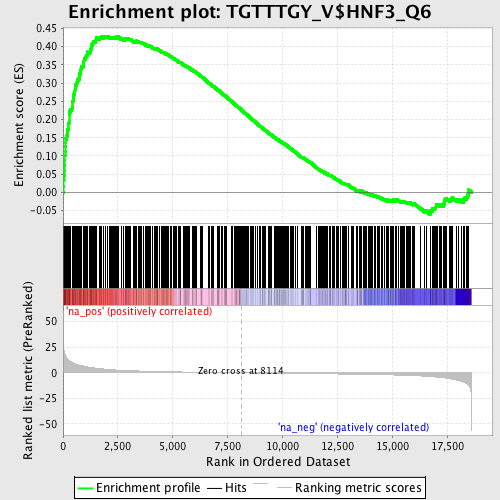
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | TGTTTGY_V$HNF3_Q6 |
| Enrichment Score (ES) | 0.42905584 |
| Normalized Enrichment Score (NES) | 1.6752486 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.056749426 |
| FWER p-Value | 0.53 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | ROBO3 | 27 | 24.638 | 0.0162 | Yes | ||
| 2 | MGAT4B | 39 | 22.458 | 0.0318 | Yes | ||
| 3 | PPP1R9B | 41 | 21.764 | 0.0474 | Yes | ||
| 4 | SERPINF1 | 55 | 20.063 | 0.0611 | Yes | ||
| 5 | TCF7L2 | 59 | 19.757 | 0.0752 | Yes | ||
| 6 | MYLK | 70 | 19.287 | 0.0885 | Yes | ||
| 7 | CA3 | 82 | 18.400 | 0.1011 | Yes | ||
| 8 | EGFL7 | 93 | 17.621 | 0.1133 | Yes | ||
| 9 | PTPN22 | 98 | 17.352 | 0.1255 | Yes | ||
| 10 | ESR1 | 111 | 16.546 | 0.1368 | Yes | ||
| 11 | SGK | 116 | 16.366 | 0.1483 | Yes | ||
| 12 | BCL9L | 173 | 14.167 | 0.1554 | Yes | ||
| 13 | ITM2B | 190 | 13.683 | 0.1644 | Yes | ||
| 14 | ROBO4 | 201 | 13.440 | 0.1735 | Yes | ||
| 15 | GRB7 | 241 | 12.611 | 0.1804 | Yes | ||
| 16 | BLNK | 244 | 12.553 | 0.1894 | Yes | ||
| 17 | MEF2C | 271 | 12.059 | 0.1966 | Yes | ||
| 18 | BCL6 | 274 | 12.030 | 0.2052 | Yes | ||
| 19 | LZTS2 | 275 | 12.026 | 0.2138 | Yes | ||
| 20 | ITPKB | 289 | 11.829 | 0.2216 | Yes | ||
| 21 | CYR61 | 321 | 11.284 | 0.2280 | Yes | ||
| 22 | ERG | 405 | 10.291 | 0.2308 | Yes | ||
| 23 | IL16 | 430 | 10.009 | 0.2367 | Yes | ||
| 24 | SOX4 | 438 | 9.925 | 0.2435 | Yes | ||
| 25 | MEIS1 | 440 | 9.904 | 0.2505 | Yes | ||
| 26 | H2AFV | 455 | 9.765 | 0.2568 | Yes | ||
| 27 | TLE4 | 473 | 9.560 | 0.2627 | Yes | ||
| 28 | DUSP6 | 481 | 9.497 | 0.2692 | Yes | ||
| 29 | PDE4D | 515 | 9.182 | 0.2740 | Yes | ||
| 30 | H1F0 | 540 | 8.900 | 0.2791 | Yes | ||
| 31 | ACVR1 | 542 | 8.880 | 0.2854 | Yes | ||
| 32 | F9 | 579 | 8.528 | 0.2895 | Yes | ||
| 33 | UBASH3A | 583 | 8.495 | 0.2955 | Yes | ||
| 34 | MGLL | 605 | 8.330 | 0.3003 | Yes | ||
| 35 | SP4 | 635 | 8.130 | 0.3046 | Yes | ||
| 36 | PDLIM2 | 663 | 7.927 | 0.3088 | Yes | ||
| 37 | PTGER2 | 708 | 7.656 | 0.3119 | Yes | ||
| 38 | KLF3 | 743 | 7.475 | 0.3154 | Yes | ||
| 39 | EFNA5 | 750 | 7.452 | 0.3204 | Yes | ||
| 40 | TCF4 | 752 | 7.435 | 0.3257 | Yes | ||
| 41 | MSI2 | 786 | 7.244 | 0.3291 | Yes | ||
| 42 | DUSP1 | 797 | 7.185 | 0.3337 | Yes | ||
| 43 | NFKBIE | 811 | 7.085 | 0.3381 | Yes | ||
| 44 | BCL11A | 818 | 7.061 | 0.3428 | Yes | ||
| 45 | THRA | 838 | 6.988 | 0.3468 | Yes | ||
| 46 | DMD | 908 | 6.710 | 0.3478 | Yes | ||
| 47 | EMCN | 918 | 6.678 | 0.3522 | Yes | ||
| 48 | H2AFY | 928 | 6.625 | 0.3564 | Yes | ||
| 49 | TGFBR1 | 938 | 6.577 | 0.3607 | Yes | ||
| 50 | SMAD5 | 952 | 6.520 | 0.3646 | Yes | ||
| 51 | DSCAM | 976 | 6.359 | 0.3679 | Yes | ||
| 52 | DDX6 | 1032 | 6.123 | 0.3693 | Yes | ||
| 53 | LEMD2 | 1041 | 6.081 | 0.3733 | Yes | ||
| 54 | SLC14A1 | 1072 | 5.943 | 0.3759 | Yes | ||
| 55 | WRN | 1098 | 5.825 | 0.3787 | Yes | ||
| 56 | RBM11 | 1110 | 5.781 | 0.3822 | Yes | ||
| 57 | RAB11B | 1126 | 5.712 | 0.3855 | Yes | ||
| 58 | CTCF | 1205 | 5.422 | 0.3851 | Yes | ||
| 59 | DOCK9 | 1232 | 5.323 | 0.3875 | Yes | ||
| 60 | GBX2 | 1249 | 5.281 | 0.3904 | Yes | ||
| 61 | ADAM15 | 1259 | 5.240 | 0.3937 | Yes | ||
| 62 | LENG9 | 1275 | 5.205 | 0.3966 | Yes | ||
| 63 | TM7SF3 | 1295 | 5.118 | 0.3993 | Yes | ||
| 64 | WBP2 | 1302 | 5.107 | 0.4026 | Yes | ||
| 65 | BBX | 1313 | 5.080 | 0.4057 | Yes | ||
| 66 | ICK | 1339 | 4.991 | 0.4079 | Yes | ||
| 67 | ZDHHC14 | 1363 | 4.934 | 0.4102 | Yes | ||
| 68 | RGS3 | 1364 | 4.932 | 0.4138 | Yes | ||
| 69 | NDST2 | 1430 | 4.751 | 0.4136 | Yes | ||
| 70 | DLGAP4 | 1481 | 4.635 | 0.4142 | Yes | ||
| 71 | AKT3 | 1488 | 4.622 | 0.4172 | Yes | ||
| 72 | TLE3 | 1498 | 4.578 | 0.4200 | Yes | ||
| 73 | PIAS1 | 1518 | 4.536 | 0.4222 | Yes | ||
| 74 | KLF7 | 1532 | 4.508 | 0.4247 | Yes | ||
| 75 | RICS | 1647 | 4.195 | 0.4214 | Yes | ||
| 76 | TRPC4AP | 1677 | 4.127 | 0.4228 | Yes | ||
| 77 | ORMDL3 | 1703 | 4.067 | 0.4243 | Yes | ||
| 78 | CD19 | 1754 | 3.951 | 0.4244 | Yes | ||
| 79 | ROM1 | 1757 | 3.940 | 0.4272 | Yes | ||
| 80 | MAPK3 | 1828 | 3.801 | 0.4260 | Yes | ||
| 81 | KIF13A | 1830 | 3.795 | 0.4287 | Yes | ||
| 82 | LRMP | 1913 | 3.608 | 0.4268 | Yes | ||
| 83 | RAB5B | 1934 | 3.568 | 0.4282 | Yes | ||
| 84 | HNRPUL1 | 2010 | 3.442 | 0.4266 | Yes | ||
| 85 | RAB35 | 2011 | 3.441 | 0.4291 | Yes | ||
| 86 | EFNA1 | 2136 | 3.202 | 0.4245 | No | ||
| 87 | TERT | 2143 | 3.195 | 0.4265 | No | ||
| 88 | ZADH2 | 2195 | 3.119 | 0.4259 | No | ||
| 89 | CTDSP1 | 2267 | 3.007 | 0.4242 | No | ||
| 90 | KCNRG | 2294 | 2.944 | 0.4249 | No | ||
| 91 | MGST1 | 2337 | 2.862 | 0.4246 | No | ||
| 92 | EYA1 | 2380 | 2.802 | 0.4243 | No | ||
| 93 | PDE3B | 2403 | 2.770 | 0.4251 | No | ||
| 94 | NFIA | 2432 | 2.728 | 0.4255 | No | ||
| 95 | LDB1 | 2439 | 2.720 | 0.4271 | No | ||
| 96 | HIST1H1C | 2482 | 2.677 | 0.4267 | No | ||
| 97 | BMF | 2510 | 2.647 | 0.4271 | No | ||
| 98 | CHN2 | 2649 | 2.487 | 0.4213 | No | ||
| 99 | PACSIN3 | 2662 | 2.473 | 0.4224 | No | ||
| 100 | PDK2 | 2749 | 2.387 | 0.4194 | No | ||
| 101 | SIPA1L2 | 2760 | 2.371 | 0.4206 | No | ||
| 102 | SLC9A3R1 | 2820 | 2.311 | 0.4190 | No | ||
| 103 | THAP11 | 2821 | 2.307 | 0.4206 | No | ||
| 104 | CALD1 | 2832 | 2.299 | 0.4217 | No | ||
| 105 | KLF5 | 2885 | 2.244 | 0.4205 | No | ||
| 106 | SLC22A2 | 2895 | 2.236 | 0.4216 | No | ||
| 107 | TBXAS1 | 2924 | 2.198 | 0.4216 | No | ||
| 108 | DCT | 2960 | 2.168 | 0.4213 | No | ||
| 109 | CSF2 | 2971 | 2.160 | 0.4223 | No | ||
| 110 | CBFB | 3025 | 2.117 | 0.4209 | No | ||
| 111 | HSPB2 | 3071 | 2.078 | 0.4199 | No | ||
| 112 | CTGF | 3202 | 1.978 | 0.4141 | No | ||
| 113 | TGM4 | 3203 | 1.976 | 0.4155 | No | ||
| 114 | MAP2K3 | 3240 | 1.945 | 0.4150 | No | ||
| 115 | LTBP1 | 3312 | 1.885 | 0.4124 | No | ||
| 116 | ATP4A | 3328 | 1.872 | 0.4129 | No | ||
| 117 | TEAD4 | 3337 | 1.866 | 0.4138 | No | ||
| 118 | ARRB2 | 3340 | 1.860 | 0.4150 | No | ||
| 119 | MAB21L2 | 3350 | 1.851 | 0.4159 | No | ||
| 120 | COL2A1 | 3416 | 1.804 | 0.4136 | No | ||
| 121 | HIST1H1E | 3453 | 1.783 | 0.4129 | No | ||
| 122 | UBE2H | 3495 | 1.747 | 0.4119 | No | ||
| 123 | PLA2G1B | 3517 | 1.730 | 0.4120 | No | ||
| 124 | MGP | 3552 | 1.711 | 0.4113 | No | ||
| 125 | RFX3 | 3576 | 1.700 | 0.4113 | No | ||
| 126 | EYA2 | 3643 | 1.660 | 0.4088 | No | ||
| 127 | ELL | 3645 | 1.660 | 0.4100 | No | ||
| 128 | LPHN2 | 3735 | 1.604 | 0.4062 | No | ||
| 129 | PHF15 | 3782 | 1.567 | 0.4048 | No | ||
| 130 | VGLL1 | 3852 | 1.527 | 0.4021 | No | ||
| 131 | OGT | 3899 | 1.499 | 0.4006 | No | ||
| 132 | GAB2 | 3908 | 1.495 | 0.4013 | No | ||
| 133 | PAIP2 | 3914 | 1.493 | 0.4021 | No | ||
| 134 | HIST1H1B | 3931 | 1.479 | 0.4023 | No | ||
| 135 | ADARB2 | 3964 | 1.459 | 0.4015 | No | ||
| 136 | ELAVL4 | 3979 | 1.448 | 0.4018 | No | ||
| 137 | ITGA3 | 4085 | 1.392 | 0.3970 | No | ||
| 138 | PITRM1 | 4144 | 1.357 | 0.3948 | No | ||
| 139 | MAPRE3 | 4155 | 1.354 | 0.3952 | No | ||
| 140 | GLP2R | 4207 | 1.330 | 0.3934 | No | ||
| 141 | PITX2 | 4241 | 1.312 | 0.3925 | No | ||
| 142 | GJB4 | 4247 | 1.309 | 0.3932 | No | ||
| 143 | BPIL1 | 4282 | 1.287 | 0.3922 | No | ||
| 144 | SOX17 | 4289 | 1.282 | 0.3928 | No | ||
| 145 | PCSK2 | 4300 | 1.277 | 0.3932 | No | ||
| 146 | GABRA2 | 4302 | 1.275 | 0.3940 | No | ||
| 147 | MITF | 4405 | 1.227 | 0.3893 | No | ||
| 148 | FCHSD2 | 4493 | 1.188 | 0.3853 | No | ||
| 149 | SELENBP1 | 4512 | 1.177 | 0.3852 | No | ||
| 150 | PRX | 4548 | 1.162 | 0.3841 | No | ||
| 151 | HOXA11 | 4567 | 1.154 | 0.3839 | No | ||
| 152 | APC | 4577 | 1.152 | 0.3843 | No | ||
| 153 | ITIH1 | 4616 | 1.138 | 0.3830 | No | ||
| 154 | PUM2 | 4657 | 1.119 | 0.3816 | No | ||
| 155 | SOX5 | 4692 | 1.104 | 0.3805 | No | ||
| 156 | HOMER2 | 4699 | 1.102 | 0.3810 | No | ||
| 157 | ULK1 | 4747 | 1.086 | 0.3792 | No | ||
| 158 | AP1S2 | 4767 | 1.075 | 0.3789 | No | ||
| 159 | CD68 | 4781 | 1.068 | 0.3789 | No | ||
| 160 | HMGN2 | 4895 | 1.023 | 0.3734 | No | ||
| 161 | NEK6 | 4914 | 1.016 | 0.3732 | No | ||
| 162 | RIT2 | 4948 | 1.004 | 0.3721 | No | ||
| 163 | CITED2 | 5036 | 0.967 | 0.3680 | No | ||
| 164 | GPLD1 | 5059 | 0.958 | 0.3675 | No | ||
| 165 | HOXC6 | 5103 | 0.941 | 0.3658 | No | ||
| 166 | PPYR1 | 5160 | 0.923 | 0.3633 | No | ||
| 167 | TMOD4 | 5242 | 0.889 | 0.3595 | No | ||
| 168 | NSD1 | 5307 | 0.864 | 0.3566 | No | ||
| 169 | SLC38A4 | 5330 | 0.855 | 0.3560 | No | ||
| 170 | GUCY2C | 5333 | 0.853 | 0.3565 | No | ||
| 171 | PPP1R1B | 5351 | 0.847 | 0.3562 | No | ||
| 172 | GPR109B | 5361 | 0.843 | 0.3563 | No | ||
| 173 | TFEC | 5495 | 0.795 | 0.3495 | No | ||
| 174 | BRD3 | 5533 | 0.777 | 0.3480 | No | ||
| 175 | HNF4G | 5550 | 0.769 | 0.3477 | No | ||
| 176 | ASB4 | 5556 | 0.767 | 0.3480 | No | ||
| 177 | HAVCR1 | 5593 | 0.748 | 0.3465 | No | ||
| 178 | ZIC4 | 5597 | 0.746 | 0.3469 | No | ||
| 179 | CNTNAP4 | 5630 | 0.736 | 0.3457 | No | ||
| 180 | NEK8 | 5632 | 0.736 | 0.3461 | No | ||
| 181 | STAT5B | 5666 | 0.724 | 0.3448 | No | ||
| 182 | PNRC1 | 5690 | 0.715 | 0.3441 | No | ||
| 183 | PDE6H | 5704 | 0.712 | 0.3439 | No | ||
| 184 | POLD4 | 5721 | 0.708 | 0.3435 | No | ||
| 185 | IL18 | 5768 | 0.692 | 0.3415 | No | ||
| 186 | SPAG4L | 5876 | 0.657 | 0.3360 | No | ||
| 187 | HOXA3 | 5879 | 0.657 | 0.3364 | No | ||
| 188 | CSAD | 5897 | 0.649 | 0.3359 | No | ||
| 189 | PROC | 5952 | 0.636 | 0.3334 | No | ||
| 190 | IGFBP1 | 5984 | 0.627 | 0.3322 | No | ||
| 191 | SLC38A3 | 6011 | 0.616 | 0.3312 | No | ||
| 192 | PHEX | 6072 | 0.602 | 0.3283 | No | ||
| 193 | FBXW11 | 6082 | 0.599 | 0.3282 | No | ||
| 194 | FGL1 | 6088 | 0.597 | 0.3284 | No | ||
| 195 | IL5RA | 6264 | 0.542 | 0.3191 | No | ||
| 196 | HOXD4 | 6287 | 0.535 | 0.3183 | No | ||
| 197 | TRPS1 | 6309 | 0.529 | 0.3175 | No | ||
| 198 | CREB5 | 6316 | 0.528 | 0.3176 | No | ||
| 199 | CALCA | 6354 | 0.514 | 0.3159 | No | ||
| 200 | SLC16A2 | 6610 | 0.438 | 0.3021 | No | ||
| 201 | BAMBI | 6651 | 0.428 | 0.3002 | No | ||
| 202 | HOXA6 | 6755 | 0.395 | 0.2948 | No | ||
| 203 | PLA2G10 | 6757 | 0.395 | 0.2951 | No | ||
| 204 | PRELP | 6815 | 0.377 | 0.2922 | No | ||
| 205 | NCAM1 | 6819 | 0.376 | 0.2923 | No | ||
| 206 | NKX2-2 | 6861 | 0.365 | 0.2903 | No | ||
| 207 | ZIC5 | 6873 | 0.363 | 0.2899 | No | ||
| 208 | CPNE1 | 7032 | 0.314 | 0.2815 | No | ||
| 209 | SIM1 | 7049 | 0.309 | 0.2808 | No | ||
| 210 | GPR81 | 7054 | 0.308 | 0.2808 | No | ||
| 211 | TIMP4 | 7080 | 0.302 | 0.2796 | No | ||
| 212 | LGI2 | 7089 | 0.298 | 0.2794 | No | ||
| 213 | SPRY1 | 7102 | 0.296 | 0.2790 | No | ||
| 214 | SRPK2 | 7114 | 0.291 | 0.2786 | No | ||
| 215 | DMRT2 | 7219 | 0.263 | 0.2730 | No | ||
| 216 | AR | 7255 | 0.252 | 0.2713 | No | ||
| 217 | GSC | 7334 | 0.228 | 0.2671 | No | ||
| 218 | TSHB | 7346 | 0.225 | 0.2667 | No | ||
| 219 | FIGN | 7393 | 0.210 | 0.2643 | No | ||
| 220 | ANKRD25 | 7402 | 0.208 | 0.2640 | No | ||
| 221 | ASCL2 | 7423 | 0.201 | 0.2630 | No | ||
| 222 | ASB5 | 7449 | 0.193 | 0.2618 | No | ||
| 223 | NTN1 | 7460 | 0.190 | 0.2614 | No | ||
| 224 | CRY2 | 7465 | 0.190 | 0.2613 | No | ||
| 225 | SERPINA6 | 7693 | 0.120 | 0.2489 | No | ||
| 226 | LHX9 | 7703 | 0.116 | 0.2485 | No | ||
| 227 | ZNF462 | 7790 | 0.091 | 0.2438 | No | ||
| 228 | POU4F3 | 7856 | 0.073 | 0.2402 | No | ||
| 229 | CHRM1 | 7872 | 0.069 | 0.2395 | No | ||
| 230 | DUSP8 | 7882 | 0.066 | 0.2390 | No | ||
| 231 | SHC3 | 7883 | 0.066 | 0.2391 | No | ||
| 232 | PCDH11X | 7915 | 0.057 | 0.2374 | No | ||
| 233 | BRS3 | 7935 | 0.052 | 0.2364 | No | ||
| 234 | TFF2 | 7984 | 0.035 | 0.2338 | No | ||
| 235 | B4GALT2 | 8021 | 0.026 | 0.2318 | No | ||
| 236 | KCMF1 | 8030 | 0.022 | 0.2314 | No | ||
| 237 | SNCB | 8035 | 0.021 | 0.2312 | No | ||
| 238 | VSNL1 | 8042 | 0.018 | 0.2308 | No | ||
| 239 | MUSK | 8074 | 0.011 | 0.2291 | No | ||
| 240 | NFE2L3 | 8100 | 0.003 | 0.2278 | No | ||
| 241 | ST5 | 8127 | -0.003 | 0.2263 | No | ||
| 242 | EVA1 | 8175 | -0.017 | 0.2238 | No | ||
| 243 | FOSB | 8234 | -0.034 | 0.2206 | No | ||
| 244 | GRIK2 | 8266 | -0.044 | 0.2189 | No | ||
| 245 | IRS1 | 8331 | -0.064 | 0.2154 | No | ||
| 246 | ITPR3 | 8344 | -0.069 | 0.2148 | No | ||
| 247 | CNTN1 | 8372 | -0.075 | 0.2134 | No | ||
| 248 | ELF4 | 8385 | -0.077 | 0.2128 | No | ||
| 249 | A2M | 8429 | -0.087 | 0.2105 | No | ||
| 250 | HAPLN1 | 8558 | -0.121 | 0.2035 | No | ||
| 251 | RAD21 | 8607 | -0.132 | 0.2009 | No | ||
| 252 | PURA | 8623 | -0.136 | 0.2002 | No | ||
| 253 | ENTPD3 | 8684 | -0.151 | 0.1970 | No | ||
| 254 | NEUROD2 | 8773 | -0.174 | 0.1923 | No | ||
| 255 | ITPA | 8871 | -0.196 | 0.1871 | No | ||
| 256 | PURG | 8965 | -0.221 | 0.1821 | No | ||
| 257 | LBX1 | 8976 | -0.223 | 0.1817 | No | ||
| 258 | JAM3 | 8996 | -0.228 | 0.1808 | No | ||
| 259 | SEMA3A | 9073 | -0.244 | 0.1768 | No | ||
| 260 | RPE65 | 9081 | -0.247 | 0.1766 | No | ||
| 261 | HAND2 | 9106 | -0.253 | 0.1754 | No | ||
| 262 | SORCS3 | 9146 | -0.263 | 0.1735 | No | ||
| 263 | HOXC13 | 9157 | -0.267 | 0.1731 | No | ||
| 264 | BANP | 9184 | -0.272 | 0.1719 | No | ||
| 265 | SEMA4C | 9203 | -0.277 | 0.1711 | No | ||
| 266 | BHLHB5 | 9343 | -0.312 | 0.1636 | No | ||
| 267 | MAP2K7 | 9344 | -0.312 | 0.1639 | No | ||
| 268 | RFX4 | 9375 | -0.320 | 0.1624 | No | ||
| 269 | IRX6 | 9398 | -0.327 | 0.1615 | No | ||
| 270 | NEUROD1 | 9417 | -0.330 | 0.1607 | No | ||
| 271 | HOXA10 | 9464 | -0.340 | 0.1584 | No | ||
| 272 | TYRO3 | 9483 | -0.345 | 0.1577 | No | ||
| 273 | SLC6A1 | 9489 | -0.346 | 0.1576 | No | ||
| 274 | JPH4 | 9498 | -0.347 | 0.1575 | No | ||
| 275 | TJP1 | 9616 | -0.375 | 0.1513 | No | ||
| 276 | STAG2 | 9638 | -0.380 | 0.1504 | No | ||
| 277 | TAGLN2 | 9642 | -0.381 | 0.1505 | No | ||
| 278 | GABRE | 9673 | -0.390 | 0.1491 | No | ||
| 279 | RNF44 | 9702 | -0.395 | 0.1479 | No | ||
| 280 | PEX16 | 9711 | -0.398 | 0.1477 | No | ||
| 281 | PRSS2 | 9754 | -0.411 | 0.1457 | No | ||
| 282 | RNF146 | 9784 | -0.419 | 0.1444 | No | ||
| 283 | NOS3 | 9790 | -0.421 | 0.1444 | No | ||
| 284 | GPR37 | 9797 | -0.424 | 0.1444 | No | ||
| 285 | DSC1 | 9854 | -0.438 | 0.1416 | No | ||
| 286 | HTR1B | 9869 | -0.442 | 0.1412 | No | ||
| 287 | ERBB3 | 9870 | -0.443 | 0.1415 | No | ||
| 288 | SFTPB | 9912 | -0.455 | 0.1395 | No | ||
| 289 | CYP7A1 | 9935 | -0.461 | 0.1387 | No | ||
| 290 | GP2 | 9962 | -0.466 | 0.1376 | No | ||
| 291 | EDN2 | 9998 | -0.477 | 0.1360 | No | ||
| 292 | OSR2 | 10004 | -0.478 | 0.1360 | No | ||
| 293 | CREB3L1 | 10029 | -0.485 | 0.1351 | No | ||
| 294 | EBF2 | 10052 | -0.493 | 0.1342 | No | ||
| 295 | GRID2 | 10073 | -0.497 | 0.1335 | No | ||
| 296 | PFTK1 | 10103 | -0.504 | 0.1322 | No | ||
| 297 | G6PC | 10136 | -0.511 | 0.1308 | No | ||
| 298 | JAG1 | 10152 | -0.514 | 0.1304 | No | ||
| 299 | UBE2B | 10160 | -0.516 | 0.1304 | No | ||
| 300 | NR0B2 | 10212 | -0.531 | 0.1279 | No | ||
| 301 | PHOX2B | 10261 | -0.547 | 0.1257 | No | ||
| 302 | SLC25A16 | 10291 | -0.555 | 0.1245 | No | ||
| 303 | RAB5A | 10363 | -0.572 | 0.1210 | No | ||
| 304 | OTX2 | 10429 | -0.588 | 0.1178 | No | ||
| 305 | VAX1 | 10464 | -0.595 | 0.1163 | No | ||
| 306 | CITED1 | 10518 | -0.607 | 0.1139 | No | ||
| 307 | CLDN2 | 10602 | -0.628 | 0.1097 | No | ||
| 308 | SNCG | 10702 | -0.648 | 0.1047 | No | ||
| 309 | JARID2 | 10856 | -0.686 | 0.0968 | No | ||
| 310 | MAPK10 | 10886 | -0.694 | 0.0957 | No | ||
| 311 | FOXP1 | 10892 | -0.694 | 0.0959 | No | ||
| 312 | OMD | 10910 | -0.699 | 0.0955 | No | ||
| 313 | CHRDL1 | 10917 | -0.699 | 0.0957 | No | ||
| 314 | PDGFRA | 10963 | -0.708 | 0.0937 | No | ||
| 315 | MAB21L1 | 10964 | -0.708 | 0.0942 | No | ||
| 316 | NFATC4 | 10972 | -0.710 | 0.0943 | No | ||
| 317 | GRIK1 | 11031 | -0.723 | 0.0916 | No | ||
| 318 | ZIC1 | 11113 | -0.741 | 0.0877 | No | ||
| 319 | ATOH8 | 11123 | -0.744 | 0.0877 | No | ||
| 320 | HCRTR2 | 11179 | -0.760 | 0.0853 | No | ||
| 321 | EGR2 | 11225 | -0.769 | 0.0833 | No | ||
| 322 | FABP1 | 11258 | -0.777 | 0.0821 | No | ||
| 323 | TNMD | 11273 | -0.781 | 0.0819 | No | ||
| 324 | OAT | 11277 | -0.781 | 0.0823 | No | ||
| 325 | KCNJ2 | 11550 | -0.854 | 0.0679 | No | ||
| 326 | RBP3 | 11648 | -0.879 | 0.0632 | No | ||
| 327 | CER1 | 11676 | -0.884 | 0.0623 | No | ||
| 328 | CACNA2D3 | 11698 | -0.891 | 0.0618 | No | ||
| 329 | ASPN | 11748 | -0.903 | 0.0598 | No | ||
| 330 | NDUFB8 | 11754 | -0.904 | 0.0601 | No | ||
| 331 | HABP2 | 11820 | -0.922 | 0.0572 | No | ||
| 332 | TEAD1 | 11838 | -0.926 | 0.0569 | No | ||
| 333 | RNF149 | 11873 | -0.936 | 0.0557 | No | ||
| 334 | DUSP10 | 11933 | -0.952 | 0.0532 | No | ||
| 335 | PTPRG | 11937 | -0.953 | 0.0537 | No | ||
| 336 | WNT5A | 11944 | -0.956 | 0.0541 | No | ||
| 337 | PTPRR | 12002 | -0.970 | 0.0516 | No | ||
| 338 | GPRC5C | 12030 | -0.980 | 0.0508 | No | ||
| 339 | B3GALT2 | 12064 | -0.987 | 0.0497 | No | ||
| 340 | MASP1 | 12134 | -1.008 | 0.0466 | No | ||
| 341 | RTN1 | 12149 | -1.012 | 0.0466 | No | ||
| 342 | SLIT3 | 12172 | -1.018 | 0.0461 | No | ||
| 343 | MMRN2 | 12181 | -1.020 | 0.0464 | No | ||
| 344 | FLRT1 | 12194 | -1.023 | 0.0465 | No | ||
| 345 | ATOH1 | 12260 | -1.040 | 0.0436 | No | ||
| 346 | GOLGB1 | 12284 | -1.047 | 0.0431 | No | ||
| 347 | JUNB | 12334 | -1.059 | 0.0412 | No | ||
| 348 | NRXN3 | 12368 | -1.068 | 0.0401 | No | ||
| 349 | DHX29 | 12450 | -1.096 | 0.0364 | No | ||
| 350 | CNTNAP2 | 12499 | -1.108 | 0.0346 | No | ||
| 351 | CRABP2 | 12532 | -1.117 | 0.0336 | No | ||
| 352 | CPEB3 | 12560 | -1.126 | 0.0330 | No | ||
| 353 | SMOC1 | 12659 | -1.159 | 0.0284 | No | ||
| 354 | NKD1 | 12723 | -1.178 | 0.0258 | No | ||
| 355 | REPS2 | 12751 | -1.186 | 0.0251 | No | ||
| 356 | IL4 | 12761 | -1.188 | 0.0255 | No | ||
| 357 | SLC6A14 | 12805 | -1.199 | 0.0240 | No | ||
| 358 | POU4F1 | 12866 | -1.221 | 0.0215 | No | ||
| 359 | NTN4 | 12881 | -1.224 | 0.0216 | No | ||
| 360 | SEMA3C | 12889 | -1.226 | 0.0221 | No | ||
| 361 | FGA | 12891 | -1.227 | 0.0230 | No | ||
| 362 | IGFALS | 12924 | -1.237 | 0.0221 | No | ||
| 363 | NR2F2 | 12936 | -1.241 | 0.0224 | No | ||
| 364 | GPM6A | 12986 | -1.258 | 0.0206 | No | ||
| 365 | ZHX2 | 13005 | -1.264 | 0.0205 | No | ||
| 366 | HNF4A | 13140 | -1.303 | 0.0140 | No | ||
| 367 | NRG1 | 13205 | -1.321 | 0.0115 | No | ||
| 368 | BCL11B | 13246 | -1.337 | 0.0102 | No | ||
| 369 | RERE | 13356 | -1.375 | 0.0052 | No | ||
| 370 | ZCCHC5 | 13394 | -1.386 | 0.0042 | No | ||
| 371 | TRIM37 | 13414 | -1.394 | 0.0041 | No | ||
| 372 | TAF15 | 13430 | -1.398 | 0.0043 | No | ||
| 373 | ANGPTL1 | 13437 | -1.401 | 0.0050 | No | ||
| 374 | SESN3 | 13502 | -1.425 | 0.0025 | No | ||
| 375 | KLF12 | 13523 | -1.432 | 0.0024 | No | ||
| 376 | GLULD1 | 13561 | -1.445 | 0.0014 | No | ||
| 377 | PIK4CB | 13566 | -1.447 | 0.0022 | No | ||
| 378 | DCI | 13574 | -1.449 | 0.0029 | No | ||
| 379 | BTG1 | 13585 | -1.454 | 0.0034 | No | ||
| 380 | DCTN1 | 13589 | -1.455 | 0.0042 | No | ||
| 381 | GTF3C1 | 13603 | -1.460 | 0.0046 | No | ||
| 382 | BAI2 | 13669 | -1.478 | 0.0021 | No | ||
| 383 | SCG3 | 13747 | -1.506 | -0.0011 | No | ||
| 384 | SDK2 | 13751 | -1.506 | -0.0002 | No | ||
| 385 | TAPBP | 13772 | -1.514 | -0.0002 | No | ||
| 386 | FOXP2 | 13796 | -1.525 | -0.0004 | No | ||
| 387 | OMG | 13841 | -1.543 | -0.0017 | No | ||
| 388 | SNAP25 | 13920 | -1.574 | -0.0049 | No | ||
| 389 | CSF3 | 13921 | -1.574 | -0.0037 | No | ||
| 390 | BRCA1 | 13970 | -1.594 | -0.0052 | No | ||
| 391 | TMOD3 | 14027 | -1.613 | -0.0072 | No | ||
| 392 | EPS8L2 | 14028 | -1.613 | -0.0060 | No | ||
| 393 | TRIM44 | 14061 | -1.624 | -0.0066 | No | ||
| 394 | EHF | 14062 | -1.624 | -0.0054 | No | ||
| 395 | TCF21 | 14119 | -1.648 | -0.0073 | No | ||
| 396 | SLC29A1 | 14179 | -1.671 | -0.0094 | No | ||
| 397 | SOX2 | 14185 | -1.673 | -0.0085 | No | ||
| 398 | BRPF1 | 14212 | -1.682 | -0.0087 | No | ||
| 399 | FGF20 | 14237 | -1.693 | -0.0088 | No | ||
| 400 | AHSG | 14329 | -1.731 | -0.0126 | No | ||
| 401 | GPR1 | 14339 | -1.735 | -0.0118 | No | ||
| 402 | KCNIP4 | 14368 | -1.750 | -0.0121 | No | ||
| 403 | CDH6 | 14424 | -1.774 | -0.0139 | No | ||
| 404 | JPH1 | 14509 | -1.814 | -0.0172 | No | ||
| 405 | SFTPC | 14513 | -1.816 | -0.0160 | No | ||
| 406 | EPHA7 | 14572 | -1.843 | -0.0179 | No | ||
| 407 | SPAG8 | 14626 | -1.866 | -0.0195 | No | ||
| 408 | FGF14 | 14660 | -1.883 | -0.0200 | No | ||
| 409 | ADAMTS10 | 14724 | -1.910 | -0.0221 | No | ||
| 410 | PRDM1 | 14786 | -1.938 | -0.0240 | No | ||
| 411 | TLL1 | 14812 | -1.953 | -0.0240 | No | ||
| 412 | ALOXE3 | 14813 | -1.953 | -0.0226 | No | ||
| 413 | AKT2 | 14825 | -1.957 | -0.0218 | No | ||
| 414 | ELF5 | 14834 | -1.962 | -0.0208 | No | ||
| 415 | A2BP1 | 14906 | -2.002 | -0.0233 | No | ||
| 416 | OSBPL2 | 14908 | -2.004 | -0.0219 | No | ||
| 417 | DHX30 | 14921 | -2.008 | -0.0211 | No | ||
| 418 | KIRREL3 | 14981 | -2.038 | -0.0229 | No | ||
| 419 | BRE | 15008 | -2.050 | -0.0229 | No | ||
| 420 | IRX5 | 15030 | -2.063 | -0.0226 | No | ||
| 421 | ALDH1A2 | 15038 | -2.069 | -0.0215 | No | ||
| 422 | ATP2C1 | 15042 | -2.070 | -0.0201 | No | ||
| 423 | RBKS | 15130 | -2.119 | -0.0234 | No | ||
| 424 | CCBL1 | 15140 | -2.128 | -0.0224 | No | ||
| 425 | GABRB1 | 15147 | -2.132 | -0.0212 | No | ||
| 426 | PAICS | 15163 | -2.142 | -0.0205 | No | ||
| 427 | NR4A3 | 15188 | -2.149 | -0.0202 | No | ||
| 428 | OMP | 15212 | -2.163 | -0.0199 | No | ||
| 429 | GTF2A1 | 15283 | -2.205 | -0.0222 | No | ||
| 430 | HOXB3 | 15359 | -2.260 | -0.0247 | No | ||
| 431 | PDYN | 15398 | -2.278 | -0.0252 | No | ||
| 432 | SLC12A1 | 15411 | -2.290 | -0.0242 | No | ||
| 433 | PIP5K2B | 15450 | -2.318 | -0.0246 | No | ||
| 434 | HR | 15521 | -2.365 | -0.0268 | No | ||
| 435 | MATN4 | 15548 | -2.383 | -0.0265 | No | ||
| 436 | GATA6 | 15560 | -2.390 | -0.0254 | No | ||
| 437 | MASP2 | 15632 | -2.441 | -0.0276 | No | ||
| 438 | GPR150 | 15659 | -2.462 | -0.0272 | No | ||
| 439 | PALMD | 15701 | -2.489 | -0.0277 | No | ||
| 440 | SEMA6A | 15754 | -2.541 | -0.0287 | No | ||
| 441 | HSPC171 | 15778 | -2.565 | -0.0282 | No | ||
| 442 | FERD3L | 15820 | -2.612 | -0.0285 | No | ||
| 443 | SVIL | 15933 | -2.716 | -0.0328 | No | ||
| 444 | FOXA2 | 15939 | -2.724 | -0.0311 | No | ||
| 445 | MXI1 | 15983 | -2.762 | -0.0315 | No | ||
| 446 | ATP2A2 | 16007 | -2.790 | -0.0307 | No | ||
| 447 | RNMT | 16310 | -3.078 | -0.0452 | No | ||
| 448 | RAD23A | 16483 | -3.284 | -0.0523 | No | ||
| 449 | DTX2 | 16489 | -3.293 | -0.0502 | No | ||
| 450 | HIP1R | 16544 | -3.365 | -0.0508 | No | ||
| 451 | PPP2R2A | 16726 | -3.640 | -0.0581 | No | ||
| 452 | EML1 | 16733 | -3.655 | -0.0558 | No | ||
| 453 | HOXC4 | 16736 | -3.657 | -0.0533 | No | ||
| 454 | PCBP1 | 16747 | -3.671 | -0.0512 | No | ||
| 455 | TPMT | 16754 | -3.675 | -0.0489 | No | ||
| 456 | NR6A1 | 16824 | -3.805 | -0.0500 | No | ||
| 457 | BCAS1 | 16825 | -3.806 | -0.0472 | No | ||
| 458 | ENTPD7 | 16832 | -3.816 | -0.0448 | No | ||
| 459 | PDCD4 | 16894 | -3.910 | -0.0454 | No | ||
| 460 | PAFAH1B3 | 16945 | -3.988 | -0.0453 | No | ||
| 461 | NR1D1 | 16950 | -4.007 | -0.0426 | No | ||
| 462 | SLC39A7 | 16960 | -4.026 | -0.0402 | No | ||
| 463 | PSMA1 | 16995 | -4.092 | -0.0391 | No | ||
| 464 | PRKAG1 | 16997 | -4.097 | -0.0362 | No | ||
| 465 | NCDN | 17016 | -4.130 | -0.0342 | No | ||
| 466 | SEMA5B | 17065 | -4.237 | -0.0338 | No | ||
| 467 | ACIN1 | 17150 | -4.422 | -0.0353 | No | ||
| 468 | YWHAG | 17186 | -4.508 | -0.0340 | No | ||
| 469 | SFRS6 | 17254 | -4.671 | -0.0343 | No | ||
| 470 | LUC7L | 17327 | -4.840 | -0.0348 | No | ||
| 471 | SMAD6 | 17333 | -4.861 | -0.0316 | No | ||
| 472 | RXRB | 17369 | -4.966 | -0.0299 | No | ||
| 473 | GNPAT | 17375 | -4.977 | -0.0266 | No | ||
| 474 | CDC25A | 17396 | -5.014 | -0.0241 | No | ||
| 475 | GADD45B | 17402 | -5.031 | -0.0208 | No | ||
| 476 | RPS6KB1 | 17420 | -5.068 | -0.0181 | No | ||
| 477 | TOB1 | 17459 | -5.211 | -0.0164 | No | ||
| 478 | E2F1 | 17613 | -5.669 | -0.0208 | No | ||
| 479 | BLOC1S2 | 17671 | -5.883 | -0.0197 | No | ||
| 480 | GEMIN4 | 17708 | -6.053 | -0.0173 | No | ||
| 481 | HESX1 | 17747 | -6.206 | -0.0150 | No | ||
| 482 | HDGF | 17927 | -7.010 | -0.0198 | No | ||
| 483 | ARHGEF6 | 18011 | -7.451 | -0.0190 | No | ||
| 484 | BZW2 | 18154 | -8.236 | -0.0209 | No | ||
| 485 | TPI1 | 18268 | -9.130 | -0.0206 | No | ||
| 486 | FSTL1 | 18298 | -9.456 | -0.0154 | No | ||
| 487 | ABCF2 | 18400 | -10.546 | -0.0134 | No | ||
| 488 | CPN1 | 18427 | -10.870 | -0.0070 | No | ||
| 489 | EPN2 | 18472 | -11.703 | -0.0010 | No | ||
| 490 | PPAT | 18495 | -12.319 | 0.0067 | No |