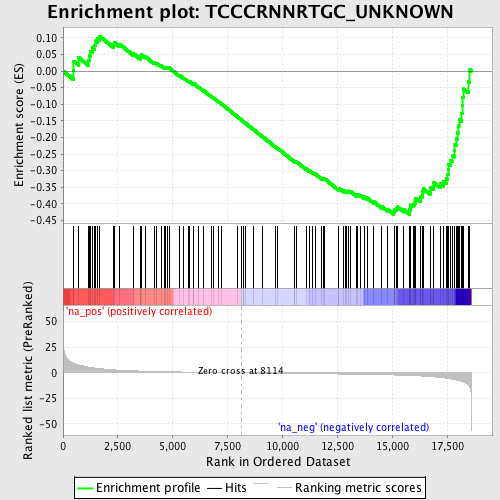
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | TCCCRNNRTGC_UNKNOWN |
| Enrichment Score (ES) | -0.43232977 |
| Normalized Enrichment Score (NES) | -1.5331867 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09347638 |
| FWER p-Value | 0.896 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | TLE4 | 473 | 9.560 | 0.0027 | No | ||
| 2 | INVS | 486 | 9.441 | 0.0299 | No | ||
| 3 | TCTA | 713 | 7.642 | 0.0402 | No | ||
| 4 | RAB1A | 1167 | 5.520 | 0.0321 | No | ||
| 5 | SNX17 | 1196 | 5.430 | 0.0466 | No | ||
| 6 | MBP | 1234 | 5.321 | 0.0603 | No | ||
| 7 | SLC4A2 | 1321 | 5.045 | 0.0705 | No | ||
| 8 | VCL | 1435 | 4.727 | 0.0784 | No | ||
| 9 | NAP1L3 | 1463 | 4.685 | 0.0908 | No | ||
| 10 | IRF2BP1 | 1571 | 4.408 | 0.0980 | No | ||
| 11 | PNRC2 | 1662 | 4.160 | 0.1054 | No | ||
| 12 | ZCCHC8 | 2289 | 2.953 | 0.0803 | No | ||
| 13 | USP15 | 2343 | 2.855 | 0.0859 | No | ||
| 14 | POLK | 2584 | 2.567 | 0.0805 | No | ||
| 15 | COX4I1 | 3217 | 1.960 | 0.0521 | No | ||
| 16 | PLAGL2 | 3522 | 1.729 | 0.0408 | No | ||
| 17 | CDK5 | 3530 | 1.721 | 0.0455 | No | ||
| 18 | MRPL24 | 3553 | 1.710 | 0.0493 | No | ||
| 19 | RHOA | 3738 | 1.602 | 0.0441 | No | ||
| 20 | CHD4 | 4159 | 1.353 | 0.0254 | No | ||
| 21 | KLK9 | 4257 | 1.305 | 0.0240 | No | ||
| 22 | NUDT13 | 4463 | 1.203 | 0.0165 | No | ||
| 23 | POFUT1 | 4635 | 1.128 | 0.0106 | No | ||
| 24 | PCTK1 | 4668 | 1.113 | 0.0122 | No | ||
| 25 | PHACTR3 | 4770 | 1.074 | 0.0099 | No | ||
| 26 | CNOT4 | 4832 | 1.050 | 0.0097 | No | ||
| 27 | NSD1 | 5307 | 0.864 | -0.0134 | No | ||
| 28 | SNX11 | 5498 | 0.795 | -0.0213 | No | ||
| 29 | RPL7 | 5703 | 0.712 | -0.0302 | No | ||
| 30 | BUB1B | 5777 | 0.690 | -0.0322 | No | ||
| 31 | INSRR | 5939 | 0.638 | -0.0390 | No | ||
| 32 | KIF11 | 5948 | 0.636 | -0.0375 | No | ||
| 33 | RPS27A | 6182 | 0.569 | -0.0484 | No | ||
| 34 | WTAP | 6419 | 0.494 | -0.0597 | No | ||
| 35 | TUBGCP3 | 6743 | 0.398 | -0.0760 | No | ||
| 36 | ZIC5 | 6873 | 0.363 | -0.0819 | No | ||
| 37 | TIMP4 | 7080 | 0.302 | -0.0922 | No | ||
| 38 | BET1 | 7225 | 0.262 | -0.0992 | No | ||
| 39 | BRS3 | 7935 | 0.052 | -0.1373 | No | ||
| 40 | TRAF3IP1 | 8151 | -0.010 | -0.1489 | No | ||
| 41 | FBS1 | 8217 | -0.028 | -0.1524 | No | ||
| 42 | FYCO1 | 8225 | -0.031 | -0.1526 | No | ||
| 43 | EYA4 | 8299 | -0.054 | -0.1564 | No | ||
| 44 | BSN | 8666 | -0.147 | -0.1758 | No | ||
| 45 | BRMS1 | 9104 | -0.252 | -0.1987 | No | ||
| 46 | MOSPD2 | 9694 | -0.394 | -0.2293 | No | ||
| 47 | NOS3 | 9790 | -0.421 | -0.2332 | No | ||
| 48 | STMN1 | 10556 | -0.615 | -0.2727 | No | ||
| 49 | COL4A3BP | 10561 | -0.617 | -0.2711 | No | ||
| 50 | HDAC8 | 10616 | -0.631 | -0.2722 | No | ||
| 51 | TCEB1 | 11078 | -0.734 | -0.2949 | No | ||
| 52 | SFRS1 | 11226 | -0.769 | -0.3006 | No | ||
| 53 | DQX1 | 11373 | -0.806 | -0.3061 | No | ||
| 54 | NEK2 | 11488 | -0.839 | -0.3098 | No | ||
| 55 | MORF4L1 | 11795 | -0.914 | -0.3237 | No | ||
| 56 | PRCC | 11858 | -0.932 | -0.3242 | No | ||
| 57 | TRIM23 | 11893 | -0.942 | -0.3233 | No | ||
| 58 | VKORC1 | 12561 | -1.126 | -0.3560 | No | ||
| 59 | CKAP4 | 12572 | -1.132 | -0.3532 | No | ||
| 60 | PLK1 | 12786 | -1.193 | -0.3612 | No | ||
| 61 | GRIN2B | 12859 | -1.218 | -0.3615 | No | ||
| 62 | LZTR1 | 12937 | -1.241 | -0.3620 | No | ||
| 63 | RB1CC1 | 12996 | -1.261 | -0.3614 | No | ||
| 64 | SNX16 | 13076 | -1.283 | -0.3619 | No | ||
| 65 | PES1 | 13361 | -1.377 | -0.3732 | No | ||
| 66 | RHOQ | 13408 | -1.392 | -0.3716 | No | ||
| 67 | ASXL2 | 13545 | -1.438 | -0.3747 | No | ||
| 68 | MAG | 13715 | -1.494 | -0.3794 | No | ||
| 69 | CASC3 | 13850 | -1.551 | -0.3821 | No | ||
| 70 | CHCHD3 | 14158 | -1.662 | -0.3937 | No | ||
| 71 | GSK3A | 14523 | -1.822 | -0.4080 | No | ||
| 72 | RPL30 | 14798 | -1.947 | -0.4171 | No | ||
| 73 | RDH10 | 15081 | -2.088 | -0.4262 | Yes | ||
| 74 | UBL3 | 15100 | -2.103 | -0.4209 | Yes | ||
| 75 | SON | 15122 | -2.116 | -0.4158 | Yes | ||
| 76 | SFRS9 | 15184 | -2.148 | -0.4128 | Yes | ||
| 77 | PCM1 | 15244 | -2.183 | -0.4095 | Yes | ||
| 78 | IPO7 | 15529 | -2.373 | -0.4179 | Yes | ||
| 79 | NUTF2 | 15794 | -2.584 | -0.4245 | Yes | ||
| 80 | PUM1 | 15797 | -2.592 | -0.4170 | Yes | ||
| 81 | VAPA | 15839 | -2.623 | -0.4114 | Yes | ||
| 82 | FUK | 15841 | -2.627 | -0.4037 | Yes | ||
| 83 | RBM6 | 15953 | -2.738 | -0.4016 | Yes | ||
| 84 | RAD50 | 16004 | -2.788 | -0.3961 | Yes | ||
| 85 | LIG1 | 16050 | -2.827 | -0.3902 | Yes | ||
| 86 | RASSF6 | 16073 | -2.844 | -0.3830 | Yes | ||
| 87 | PSMD11 | 16285 | -3.054 | -0.3854 | Yes | ||
| 88 | CSNK2A1 | 16302 | -3.072 | -0.3772 | Yes | ||
| 89 | TTC1 | 16364 | -3.129 | -0.3712 | Yes | ||
| 90 | ISL1 | 16376 | -3.139 | -0.3625 | Yes | ||
| 91 | ORC4L | 16414 | -3.183 | -0.3551 | Yes | ||
| 92 | NMT1 | 16732 | -3.654 | -0.3615 | Yes | ||
| 93 | ACADVL | 16735 | -3.656 | -0.3508 | Yes | ||
| 94 | SMPD3 | 16878 | -3.890 | -0.3470 | Yes | ||
| 95 | ABCB6 | 16893 | -3.909 | -0.3362 | Yes | ||
| 96 | RRS1 | 17190 | -4.513 | -0.3389 | Yes | ||
| 97 | PPP1R8 | 17334 | -4.861 | -0.3322 | Yes | ||
| 98 | RPP30 | 17466 | -5.232 | -0.3239 | Yes | ||
| 99 | MIR16 | 17537 | -5.464 | -0.3115 | Yes | ||
| 100 | USP16 | 17559 | -5.514 | -0.2964 | Yes | ||
| 101 | UBADC1 | 17572 | -5.558 | -0.2806 | Yes | ||
| 102 | MRPL50 | 17668 | -5.868 | -0.2684 | Yes | ||
| 103 | GSPT1 | 17741 | -6.175 | -0.2541 | Yes | ||
| 104 | PA2G4 | 17835 | -6.589 | -0.2397 | Yes | ||
| 105 | GCN5L2 | 17859 | -6.653 | -0.2213 | Yes | ||
| 106 | KPNB1 | 17917 | -6.958 | -0.2038 | Yes | ||
| 107 | HSPH1 | 17959 | -7.187 | -0.1848 | Yes | ||
| 108 | PMM2 | 18003 | -7.395 | -0.1653 | Yes | ||
| 109 | SCAMP4 | 18045 | -7.670 | -0.1448 | Yes | ||
| 110 | IFRD2 | 18160 | -8.304 | -0.1265 | Yes | ||
| 111 | EIF5A | 18184 | -8.462 | -0.1027 | Yes | ||
| 112 | XPOT | 18212 | -8.637 | -0.0787 | Yes | ||
| 113 | GART | 18243 | -8.950 | -0.0539 | Yes | ||
| 114 | EIF2B4 | 18461 | -11.400 | -0.0320 | Yes | ||
| 115 | POLR1A | 18533 | -13.643 | 0.0045 | Yes |