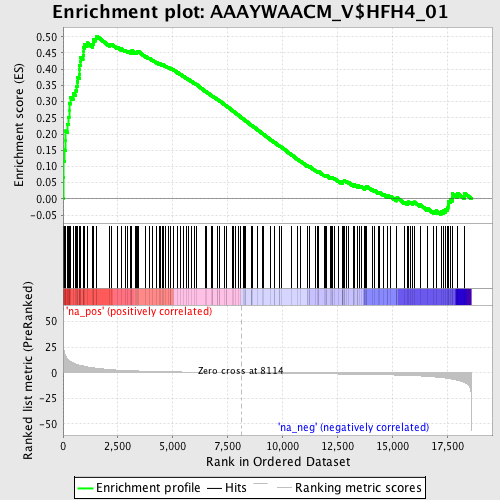
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set04_transDMproB_versus_transDMpreB |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | AAAYWAACM_V$HFH4_01 |
| Enrichment Score (ES) | 0.50205255 |
| Normalized Enrichment Score (NES) | 1.785759 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.037565187 |
| FWER p-Value | 0.175 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | BOK | 8 | 34.984 | 0.0650 | Yes | ||
| 2 | IMPDH1 | 16 | 28.243 | 0.1174 | Yes | ||
| 3 | TCF7L2 | 59 | 19.757 | 0.1520 | Yes | ||
| 4 | SGK | 116 | 16.366 | 0.1796 | Yes | ||
| 5 | GFRA1 | 120 | 16.047 | 0.2094 | Yes | ||
| 6 | NDRG1 | 200 | 13.450 | 0.2303 | Yes | ||
| 7 | GRB7 | 241 | 12.611 | 0.2517 | Yes | ||
| 8 | MEF2C | 271 | 12.059 | 0.2726 | Yes | ||
| 9 | BCL6 | 274 | 12.030 | 0.2950 | Yes | ||
| 10 | CYR61 | 321 | 11.284 | 0.3136 | Yes | ||
| 11 | H2AFV | 455 | 9.765 | 0.3247 | Yes | ||
| 12 | PPARGC1A | 584 | 8.495 | 0.3336 | Yes | ||
| 13 | MGLL | 605 | 8.330 | 0.3481 | Yes | ||
| 14 | MAP3K3 | 642 | 8.089 | 0.3612 | Yes | ||
| 15 | HOXB4 | 664 | 7.921 | 0.3749 | Yes | ||
| 16 | KLF3 | 743 | 7.475 | 0.3847 | Yes | ||
| 17 | TCF4 | 752 | 7.435 | 0.3981 | Yes | ||
| 18 | NFIX | 759 | 7.390 | 0.4116 | Yes | ||
| 19 | MSI2 | 786 | 7.244 | 0.4237 | Yes | ||
| 20 | DUSP1 | 797 | 7.185 | 0.4366 | Yes | ||
| 21 | DMD | 908 | 6.710 | 0.4432 | Yes | ||
| 22 | IDH1 | 927 | 6.636 | 0.4546 | Yes | ||
| 23 | HOXA9 | 932 | 6.609 | 0.4668 | Yes | ||
| 24 | ATF7IP | 969 | 6.391 | 0.4768 | Yes | ||
| 25 | PPAP2B | 1091 | 5.848 | 0.4811 | Yes | ||
| 26 | ICK | 1339 | 4.991 | 0.4771 | Yes | ||
| 27 | PBX1 | 1383 | 4.889 | 0.4839 | Yes | ||
| 28 | TBCC | 1389 | 4.867 | 0.4927 | Yes | ||
| 29 | TLE3 | 1498 | 4.578 | 0.4954 | Yes | ||
| 30 | KLF7 | 1532 | 4.508 | 0.5021 | Yes | ||
| 31 | EFNA1 | 2136 | 3.202 | 0.4753 | No | ||
| 32 | ELOVL6 | 2206 | 3.099 | 0.4774 | No | ||
| 33 | FGF13 | 2478 | 2.679 | 0.4677 | No | ||
| 34 | CHN2 | 2649 | 2.487 | 0.4631 | No | ||
| 35 | CALD1 | 2832 | 2.299 | 0.4576 | No | ||
| 36 | DNAJB12 | 2946 | 2.180 | 0.4555 | No | ||
| 37 | ZIC3 | 3056 | 2.089 | 0.4535 | No | ||
| 38 | SNCAIP | 3131 | 2.031 | 0.4533 | No | ||
| 39 | FUT11 | 3136 | 2.029 | 0.4569 | No | ||
| 40 | IRS4 | 3283 | 1.913 | 0.4525 | No | ||
| 41 | MAB21L2 | 3350 | 1.851 | 0.4524 | No | ||
| 42 | SCRG1 | 3390 | 1.822 | 0.4537 | No | ||
| 43 | TRIM8 | 3449 | 1.785 | 0.4539 | No | ||
| 44 | ARHGEF2 | 3773 | 1.571 | 0.4393 | No | ||
| 45 | PAIP2 | 3914 | 1.493 | 0.4345 | No | ||
| 46 | ITGA3 | 4085 | 1.392 | 0.4279 | No | ||
| 47 | RGL2 | 4255 | 1.307 | 0.4212 | No | ||
| 48 | MITF | 4405 | 1.227 | 0.4154 | No | ||
| 49 | EVL | 4429 | 1.218 | 0.4164 | No | ||
| 50 | PRX | 4548 | 1.162 | 0.4122 | No | ||
| 51 | HOXA11 | 4567 | 1.154 | 0.4134 | No | ||
| 52 | CAST | 4674 | 1.112 | 0.4097 | No | ||
| 53 | CD68 | 4781 | 1.068 | 0.4060 | No | ||
| 54 | GOLGA1 | 4884 | 1.026 | 0.4024 | No | ||
| 55 | HMGN2 | 4895 | 1.023 | 0.4037 | No | ||
| 56 | MYLIP | 5023 | 0.972 | 0.3987 | No | ||
| 57 | PAQR9 | 5228 | 0.896 | 0.3893 | No | ||
| 58 | NLGN3 | 5340 | 0.850 | 0.3848 | No | ||
| 59 | TFEC | 5495 | 0.795 | 0.3780 | No | ||
| 60 | CNTNAP4 | 5630 | 0.736 | 0.3721 | No | ||
| 61 | BCOR | 5733 | 0.704 | 0.3679 | No | ||
| 62 | SOX1 | 5843 | 0.669 | 0.3632 | No | ||
| 63 | ANGPT2 | 5975 | 0.629 | 0.3573 | No | ||
| 64 | PHEX | 6072 | 0.602 | 0.3532 | No | ||
| 65 | EPHB1 | 6490 | 0.470 | 0.3315 | No | ||
| 66 | MYL1 | 6552 | 0.452 | 0.3290 | No | ||
| 67 | HOXA6 | 6755 | 0.395 | 0.3188 | No | ||
| 68 | HOXB8 | 6805 | 0.381 | 0.3169 | No | ||
| 69 | NCAM1 | 6819 | 0.376 | 0.3169 | No | ||
| 70 | CPNE1 | 7032 | 0.314 | 0.3060 | No | ||
| 71 | SIM1 | 7049 | 0.309 | 0.3057 | No | ||
| 72 | SOX3 | 7115 | 0.291 | 0.3027 | No | ||
| 73 | BMP7 | 7124 | 0.288 | 0.3028 | No | ||
| 74 | GSC | 7334 | 0.228 | 0.2919 | No | ||
| 75 | NTN1 | 7460 | 0.190 | 0.2855 | No | ||
| 76 | CRY2 | 7465 | 0.190 | 0.2856 | No | ||
| 77 | LHX9 | 7703 | 0.116 | 0.2730 | No | ||
| 78 | PIK3C2A | 7753 | 0.100 | 0.2705 | No | ||
| 79 | ITGBL1 | 7857 | 0.072 | 0.2651 | No | ||
| 80 | BUB3 | 8006 | 0.028 | 0.2571 | No | ||
| 81 | NFE2L3 | 8100 | 0.003 | 0.2520 | No | ||
| 82 | FOSB | 8234 | -0.034 | 0.2449 | No | ||
| 83 | GRIK2 | 8266 | -0.044 | 0.2433 | No | ||
| 84 | STARD13 | 8293 | -0.052 | 0.2420 | No | ||
| 85 | IRS1 | 8331 | -0.064 | 0.2401 | No | ||
| 86 | SPON1 | 8588 | -0.129 | 0.2265 | No | ||
| 87 | TBX3 | 8605 | -0.132 | 0.2258 | No | ||
| 88 | RAD21 | 8607 | -0.132 | 0.2260 | No | ||
| 89 | PURA | 8623 | -0.136 | 0.2255 | No | ||
| 90 | NUDT11 | 8875 | -0.196 | 0.2122 | No | ||
| 91 | SEMA3A | 9073 | -0.244 | 0.2020 | No | ||
| 92 | TACSTD2 | 9139 | -0.261 | 0.1990 | No | ||
| 93 | HOXA10 | 9464 | -0.340 | 0.1820 | No | ||
| 94 | TJP1 | 9616 | -0.375 | 0.1746 | No | ||
| 95 | STAG2 | 9638 | -0.380 | 0.1741 | No | ||
| 96 | HTR1B | 9869 | -0.442 | 0.1625 | No | ||
| 97 | PPP1CB | 9879 | -0.446 | 0.1628 | No | ||
| 98 | GSH1 | 9953 | -0.464 | 0.1597 | No | ||
| 99 | OTX2 | 10429 | -0.588 | 0.1351 | No | ||
| 100 | ENPP2 | 10681 | -0.645 | 0.1227 | No | ||
| 101 | LGI1 | 10823 | -0.678 | 0.1163 | No | ||
| 102 | ATOH8 | 11123 | -0.744 | 0.1015 | No | ||
| 103 | SYTL2 | 11137 | -0.749 | 0.1022 | No | ||
| 104 | EGR2 | 11225 | -0.769 | 0.0989 | No | ||
| 105 | HSF2 | 11244 | -0.772 | 0.0994 | No | ||
| 106 | CLN5 | 11491 | -0.840 | 0.0876 | No | ||
| 107 | SIN3A | 11586 | -0.863 | 0.0841 | No | ||
| 108 | RBP3 | 11648 | -0.879 | 0.0825 | No | ||
| 109 | CXXC5 | 11661 | -0.881 | 0.0835 | No | ||
| 110 | GPR85 | 11894 | -0.942 | 0.0726 | No | ||
| 111 | WNT5A | 11944 | -0.956 | 0.0718 | No | ||
| 112 | CRIM1 | 11989 | -0.967 | 0.0712 | No | ||
| 113 | C1QTNF6 | 12024 | -0.977 | 0.0712 | No | ||
| 114 | SLIT3 | 12172 | -1.018 | 0.0651 | No | ||
| 115 | GPR142 | 12221 | -1.030 | 0.0644 | No | ||
| 116 | ATOH1 | 12260 | -1.040 | 0.0643 | No | ||
| 117 | UBE3A | 12268 | -1.042 | 0.0659 | No | ||
| 118 | NRXN3 | 12368 | -1.068 | 0.0625 | No | ||
| 119 | CPEB3 | 12560 | -1.126 | 0.0543 | No | ||
| 120 | MAFF | 12717 | -1.175 | 0.0480 | No | ||
| 121 | NKD1 | 12723 | -1.178 | 0.0499 | No | ||
| 122 | PGM2 | 12735 | -1.181 | 0.0515 | No | ||
| 123 | REPS2 | 12751 | -1.186 | 0.0529 | No | ||
| 124 | FLRT3 | 12765 | -1.189 | 0.0545 | No | ||
| 125 | ACCN2 | 12803 | -1.198 | 0.0547 | No | ||
| 126 | BDNF | 12829 | -1.208 | 0.0556 | No | ||
| 127 | NR2F2 | 12936 | -1.241 | 0.0522 | No | ||
| 128 | ZHX2 | 13005 | -1.264 | 0.0509 | No | ||
| 129 | HOXC12 | 13220 | -1.328 | 0.0417 | No | ||
| 130 | BCL11B | 13246 | -1.337 | 0.0429 | No | ||
| 131 | GRIK3 | 13280 | -1.348 | 0.0436 | No | ||
| 132 | NRK | 13423 | -1.396 | 0.0385 | No | ||
| 133 | ANGPTL1 | 13437 | -1.401 | 0.0404 | No | ||
| 134 | KLF12 | 13523 | -1.432 | 0.0385 | No | ||
| 135 | DTNA | 13594 | -1.457 | 0.0374 | No | ||
| 136 | SCG3 | 13747 | -1.506 | 0.0320 | No | ||
| 137 | SDK2 | 13751 | -1.506 | 0.0347 | No | ||
| 138 | FOXP2 | 13796 | -1.525 | 0.0351 | No | ||
| 139 | MCC | 13818 | -1.536 | 0.0369 | No | ||
| 140 | OMG | 13841 | -1.543 | 0.0385 | No | ||
| 141 | ADAM11 | 14094 | -1.641 | 0.0279 | No | ||
| 142 | C1QTNF3 | 14211 | -1.682 | 0.0248 | No | ||
| 143 | SLC39A8 | 14369 | -1.750 | 0.0196 | No | ||
| 144 | CRH | 14419 | -1.771 | 0.0202 | No | ||
| 145 | STX16 | 14588 | -1.850 | 0.0146 | No | ||
| 146 | FGF17 | 14781 | -1.935 | 0.0078 | No | ||
| 147 | PRDM1 | 14786 | -1.938 | 0.0112 | No | ||
| 148 | A2BP1 | 14906 | -2.002 | 0.0085 | No | ||
| 149 | NR4A3 | 15188 | -2.149 | -0.0027 | No | ||
| 150 | NEO1 | 15207 | -2.161 | 0.0003 | No | ||
| 151 | CD36 | 15215 | -2.164 | 0.0040 | No | ||
| 152 | GATA6 | 15560 | -2.390 | -0.0102 | No | ||
| 153 | PALMD | 15701 | -2.489 | -0.0131 | No | ||
| 154 | GPX1 | 15722 | -2.510 | -0.0095 | No | ||
| 155 | CSMD3 | 15819 | -2.609 | -0.0099 | No | ||
| 156 | FOXA2 | 15939 | -2.724 | -0.0112 | No | ||
| 157 | ATP2A2 | 16007 | -2.790 | -0.0096 | No | ||
| 158 | INSM1 | 16276 | -3.048 | -0.0185 | No | ||
| 159 | P2RY10 | 16597 | -3.443 | -0.0294 | No | ||
| 160 | PDCD4 | 16894 | -3.910 | -0.0381 | No | ||
| 161 | NEDD4 | 17006 | -4.114 | -0.0364 | No | ||
| 162 | EIF4G1 | 17226 | -4.598 | -0.0397 | No | ||
| 163 | SNTB2 | 17350 | -4.899 | -0.0372 | No | ||
| 164 | RPS6KB1 | 17420 | -5.068 | -0.0315 | No | ||
| 165 | CDC2L2 | 17520 | -5.382 | -0.0268 | No | ||
| 166 | DOCK3 | 17549 | -5.495 | -0.0181 | No | ||
| 167 | ING3 | 17574 | -5.561 | -0.0090 | No | ||
| 168 | HSPA2 | 17646 | -5.787 | -0.0020 | No | ||
| 169 | IRF4 | 17737 | -6.144 | 0.0046 | No | ||
| 170 | HESX1 | 17747 | -6.206 | 0.0157 | No | ||
| 171 | NCOA5 | 17983 | -7.284 | 0.0166 | No | ||
| 172 | FSTL1 | 18298 | -9.456 | 0.0172 | No |